Gene
KWMTBOMO03698 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA002750
Annotation
PREDICTED:_pyruvate_dehydrogenase_E1_component_beta_subunit_isoform_X1_[Bombyx_mori]
Full name
Pyruvate dehydrogenase E1 component subunit beta
Location in the cell
Cytoplasmic Reliability : 1.322 Mitochondrial Reliability : 1.93
Sequence
CDS
ATGGCATTAAAATCATCGCCGGCTGTTCTCGGGATGCTCACTCGATTATCCCGTCGCAGTTTTGCCACATCGAAAGCTCTGGCTTCTAAGCCGGTGACTGTAAGAGATGCTTTAAATCAAGCGATCGACGAGGAGATGGAGAGAGACGAGAAAGTCTTCGTTTTGGGAGAGGAAGTCGCTCAATACGATGGTGCCTACAAAGTTACAAGGGGTCTATGGAAGAAATATGGTGATAAAAGAGTTATCGACACACCCATCACAGAGGCAGGATTCGCCGGTATAGCTGTTGGAGCTGCTTTTGCAGGGCTCAAGCCTATTTGTGAATTCATGACCTTTAACTTCTCTATGCAAGCTATTGATCATATAATAAACTCAGCCGCCAAGACCTTCTATATGTCAGCTGGCACAGTACCAGTGCCAATAGTATTCCGAGGACCTAATGGAGCTGCCTCCGGTGTTGCCGCACAACATTCCCAGTGCTTTGGTGCCTGGTACAGCCACTGTCCAGGCCTTAAAGTTCTAATGCCTTATTCAGCAGAGGATGCTAAAGGTCTCCTGAAAGCTGCCATCAGGGATCCCGACCCAGTTGTTATGTTGGAAGATGAAATTATGTATGGTATACCATTCCCAATGTCAGATGAAGCCCAGTCCAAAGACTTTGTACTTCCTATTGGCAAAGCTAAGGTAGAAAGGGAGGGTCGGCATATCACGCTAGTGTGCGCTGGACGAGGCACCGACACCGCGCTCAAGGCCGCTGAACAACTGGCTGGAAGCAAGGGTATAGAATGTGAAGTAGTAAATCTAAGAACAATAAGGCCAATGGATTTCGACACGATCGCTCGGTCCATTGCCAAAACACATCATCTCATTACTGTAGAACAAGGATGGCCGCAATCTGGAATCGGCGCAGAGATCTGCGCCCGCGTCATGGAGTCGCCGTCGTTCTTCGAGCTAGATGCGCCGGTGTGGCGAGTGTGCGGCGCCGACGTGCCCATGCCCTACGCCAGGACGCTCGAGGCGCACGCGGTGCCCGGACCCGCCGACGTGGTGGACGCCGTCACCAACGTGCTCGGCAACAAAATCAGTGCAGTGCAAGCCCAGTAG
Protein
MALKSSPAVLGMLTRLSRRSFATSKALASKPVTVRDALNQAIDEEMERDEKVFVLGEEVAQYDGAYKVTRGLWKKYGDKRVIDTPITEAGFAGIAVGAAFAGLKPICEFMTFNFSMQAIDHIINSAAKTFYMSAGTVPVPIVFRGPNGAASGVAAQHSQCFGAWYSHCPGLKVLMPYSAEDAKGLLKAAIRDPDPVVMLEDEIMYGIPFPMSDEAQSKDFVLPIGKAKVEREGRHITLVCAGRGTDTALKAAEQLAGSKGIECEVVNLRTIRPMDFDTIARSIAKTHHLITVEQGWPQSGIGAEICARVMESPSFFELDAPVWRVCGADVPMPYARTLEAHAVPGPADVVDAVTNVLGNKISAVQAQ
Summary
Description
The pyruvate dehydrogenase complex catalyzes the overall conversion of pyruvate to acetyl-CoA and CO2.
Catalytic Activity
[dihydrolipoyllysine-residue acetyltransferase]-(R)-N(6)-lipoyl-L-lysine + H(+) + pyruvate = [dihydrolipoyllysine-residue acetyltransferase]-(R)-N(6)-(S(8)-acetyldihydrolipoyl)-L-lysine + CO2
Cofactor
thiamine diphosphate
Uniprot
H9IZR5
A0A194R8S3
A0A2H1V0P1
S4PDP5
A0A194PX57
A0A3S2NM84
+ More
A0A212FA10 A0A182QQ36 A0A1S4H3X9 Q7QDU3 A0A336MXN1 A0A182UX13 A0A336MZK5 W5J7J0 A0A182XB08 A0A182HX81 A0A182LN80 A0A182NDJ7 A0A2M3Z685 A0A182K007 A0A182W3R4 A0A2M4AEX7 A0A2M4BSD4 B0XA87 A0A182LUG9 A0A182RBE4 A0A182YLY1 T1E2S0 Q17D51 A0A084VUL4 A0A182PEU7 A0A1S4F7M2 A0A1Q3FG45 A0A023ERL5 A0A182JDB8 A0A336MUR1 A0A1B6L2R2 A0A2P8XMD5 U5ESA2 V9IHF4 A0A2A3E2G8 A0A182FM08 Q1HPJ1 A0A1B6GEQ8 A0A1B6EG39 A0A195F310 A0A195DCA7 A0A158NE37 A0A067RJD6 A0A151WFB4 A0A195BBX2 A0A0A9Y541 E2C3B2 K7IXW5 A0A1B0EZK0 A0A1Y1LZ82 A0A2J7PL96 A0A1L8E0L6 A0A226DAT4 A0A182U880 A0A1D2N7U1 E9J7R1 U4TT33 A0A1I8Q9Y7 E2AI43 A0A1L8ECM4 A0A2Z5RE12 T1PC65 A0A1L8ED34 A0A151IGB1 A0A0V0GB25 A0A1I8MP09 Q8IGJ4 Q7K5K3 A0A1W4U638 A0A0K8TSX5 B4HZ58 B4NBF7 D6X401 A0A023FCH4 B4PQ74 A0A0L7QL62 T1IFK5 B3P5L5 A0A0N8E2A6 R4WDA4 A0A026W2Y6 A0A0K8VLY4 A0A034WCB5 A0A0K8UPM2 A0A1B0DP56 A0A154PL96 B4K754 E9HA87 A0A224XF23 W8B9P1 A0A2H8TQV1 A0A0M9A1W7 A0A2T7NTB3 A0A310SVU9
A0A212FA10 A0A182QQ36 A0A1S4H3X9 Q7QDU3 A0A336MXN1 A0A182UX13 A0A336MZK5 W5J7J0 A0A182XB08 A0A182HX81 A0A182LN80 A0A182NDJ7 A0A2M3Z685 A0A182K007 A0A182W3R4 A0A2M4AEX7 A0A2M4BSD4 B0XA87 A0A182LUG9 A0A182RBE4 A0A182YLY1 T1E2S0 Q17D51 A0A084VUL4 A0A182PEU7 A0A1S4F7M2 A0A1Q3FG45 A0A023ERL5 A0A182JDB8 A0A336MUR1 A0A1B6L2R2 A0A2P8XMD5 U5ESA2 V9IHF4 A0A2A3E2G8 A0A182FM08 Q1HPJ1 A0A1B6GEQ8 A0A1B6EG39 A0A195F310 A0A195DCA7 A0A158NE37 A0A067RJD6 A0A151WFB4 A0A195BBX2 A0A0A9Y541 E2C3B2 K7IXW5 A0A1B0EZK0 A0A1Y1LZ82 A0A2J7PL96 A0A1L8E0L6 A0A226DAT4 A0A182U880 A0A1D2N7U1 E9J7R1 U4TT33 A0A1I8Q9Y7 E2AI43 A0A1L8ECM4 A0A2Z5RE12 T1PC65 A0A1L8ED34 A0A151IGB1 A0A0V0GB25 A0A1I8MP09 Q8IGJ4 Q7K5K3 A0A1W4U638 A0A0K8TSX5 B4HZ58 B4NBF7 D6X401 A0A023FCH4 B4PQ74 A0A0L7QL62 T1IFK5 B3P5L5 A0A0N8E2A6 R4WDA4 A0A026W2Y6 A0A0K8VLY4 A0A034WCB5 A0A0K8UPM2 A0A1B0DP56 A0A154PL96 B4K754 E9HA87 A0A224XF23 W8B9P1 A0A2H8TQV1 A0A0M9A1W7 A0A2T7NTB3 A0A310SVU9
EC Number
1.2.4.1
Pubmed
19121390
26354079
23622113
22118469
12364791
20920257
+ More
23761445 20966253 25244985 24330624 17510324 24438588 24945155 29403074 21347285 24845553 25401762 26823975 20798317 20075255 28004739 30926861 27289101 21282665 23537049 26760975 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26369729 17994087 18362917 19820115 25474469 17550304 23691247 24508170 30249741 25348373 21292972 24495485
23761445 20966253 25244985 24330624 17510324 24438588 24945155 29403074 21347285 24845553 25401762 26823975 20798317 20075255 28004739 30926861 27289101 21282665 23537049 26760975 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26369729 17994087 18362917 19820115 25474469 17550304 23691247 24508170 30249741 25348373 21292972 24495485
EMBL
BABH01038020
BABH01038021
KQ460500
KPJ14238.1
ODYU01000144
SOQ34415.1
+ More
GAIX01003581 JAA88979.1 KQ459589 KPI97598.1 RSAL01000004 RVE54562.1 AGBW02009536 OWR50563.1 AXCN02001759 AAAB01008849 EAA07168.2 UFQT01002647 SSX33849.1 SSX33847.1 ADMH02002024 ETN59931.1 APCN01005148 GGFM01003207 MBW23958.1 GGFK01005867 MBW39188.1 GGFJ01006855 MBW55996.1 DS232572 EDS43584.1 AXCM01004341 GALA01001045 JAA93807.1 CH477300 EAT44269.1 ATLV01016963 KE525139 KFB41658.1 GFDL01008534 JAV26511.1 GAPW01002274 JAC11324.1 UFQT01002648 SSX33850.1 GEBQ01022037 JAT17940.1 PYGN01001729 PSN33167.1 GANO01002449 JAB57422.1 JR047731 AEY60490.1 KZ288455 PBC25522.1 DQ443411 ABF51500.1 GECZ01031345 GECZ01018906 GECZ01016442 GECZ01016074 GECZ01008852 JAS38424.1 JAS50863.1 JAS53327.1 JAS53695.1 JAS60917.1 GEDC01000400 JAS36898.1 KQ981855 KYN34771.1 KQ980989 KYN10545.1 ADTU01001291 KK852597 KDR20584.1 KQ983219 KYQ46521.1 KQ976522 KYM82066.1 GBHO01015438 GBHO01015437 GBRD01002408 GBRD01002407 GDHC01008432 GDHC01003715 JAG28166.1 JAG28167.1 JAG63413.1 JAQ10197.1 JAQ14914.1 GL452318 EFN77558.1 AJWK01022514 GEZM01048918 GEZM01048917 JAV76287.1 NEVH01024426 PNF17106.1 GFDF01001816 JAV12268.1 LNIX01000025 MH799926 OXA42665.1 QBH74085.1 LJIJ01000160 ODN01322.1 GL768602 EFZ11142.1 KB631615 ERL84684.1 GL439639 EFN66894.1 GFDG01002309 JAV16490.1 FX983733 BAX07232.1 KA645715 AFP60344.1 GFDG01002310 JAV16489.1 KQ977726 KYN00275.1 GECL01001072 JAP05052.1 BT001756 AAN71511.1 AE014297 AY047573 AAF56855.2 AAK77305.1 GDAI01000141 JAI17462.1 CH480819 EDW53315.1 CH964232 EDW81121.2 KQ971379 EEZ97343.1 GBBI01000234 JAC18478.1 CM000160 EDW98343.1 KQ414934 KOC59353.1 ACPB03005784 CH954182 EDV53265.1 GDIQ01102606 GDIP01043210 GDIQ01068820 LRGB01002127 JAM60505.1 JAN25917.1 KZS08995.1 AK417369 BAN20584.1 KK107488 QOIP01000012 EZA49951.1 RLU16080.1 GDHF01012739 JAI39575.1 GAKP01006623 JAC52329.1 GDHF01023682 GDHF01022729 JAI28632.1 JAI29585.1 AJVK01007913 AJVK01007914 KQ434954 KZC12527.1 CH933806 EDW16367.1 GL732610 EFX71388.1 GFTR01005350 JAW11076.1 GAMC01012792 GAMC01012790 JAB93763.1 GFXV01004761 MBW16566.1 KQ435794 KOX74119.1 PZQS01000009 PVD24420.1 KQ759866 OAD62493.1
GAIX01003581 JAA88979.1 KQ459589 KPI97598.1 RSAL01000004 RVE54562.1 AGBW02009536 OWR50563.1 AXCN02001759 AAAB01008849 EAA07168.2 UFQT01002647 SSX33849.1 SSX33847.1 ADMH02002024 ETN59931.1 APCN01005148 GGFM01003207 MBW23958.1 GGFK01005867 MBW39188.1 GGFJ01006855 MBW55996.1 DS232572 EDS43584.1 AXCM01004341 GALA01001045 JAA93807.1 CH477300 EAT44269.1 ATLV01016963 KE525139 KFB41658.1 GFDL01008534 JAV26511.1 GAPW01002274 JAC11324.1 UFQT01002648 SSX33850.1 GEBQ01022037 JAT17940.1 PYGN01001729 PSN33167.1 GANO01002449 JAB57422.1 JR047731 AEY60490.1 KZ288455 PBC25522.1 DQ443411 ABF51500.1 GECZ01031345 GECZ01018906 GECZ01016442 GECZ01016074 GECZ01008852 JAS38424.1 JAS50863.1 JAS53327.1 JAS53695.1 JAS60917.1 GEDC01000400 JAS36898.1 KQ981855 KYN34771.1 KQ980989 KYN10545.1 ADTU01001291 KK852597 KDR20584.1 KQ983219 KYQ46521.1 KQ976522 KYM82066.1 GBHO01015438 GBHO01015437 GBRD01002408 GBRD01002407 GDHC01008432 GDHC01003715 JAG28166.1 JAG28167.1 JAG63413.1 JAQ10197.1 JAQ14914.1 GL452318 EFN77558.1 AJWK01022514 GEZM01048918 GEZM01048917 JAV76287.1 NEVH01024426 PNF17106.1 GFDF01001816 JAV12268.1 LNIX01000025 MH799926 OXA42665.1 QBH74085.1 LJIJ01000160 ODN01322.1 GL768602 EFZ11142.1 KB631615 ERL84684.1 GL439639 EFN66894.1 GFDG01002309 JAV16490.1 FX983733 BAX07232.1 KA645715 AFP60344.1 GFDG01002310 JAV16489.1 KQ977726 KYN00275.1 GECL01001072 JAP05052.1 BT001756 AAN71511.1 AE014297 AY047573 AAF56855.2 AAK77305.1 GDAI01000141 JAI17462.1 CH480819 EDW53315.1 CH964232 EDW81121.2 KQ971379 EEZ97343.1 GBBI01000234 JAC18478.1 CM000160 EDW98343.1 KQ414934 KOC59353.1 ACPB03005784 CH954182 EDV53265.1 GDIQ01102606 GDIP01043210 GDIQ01068820 LRGB01002127 JAM60505.1 JAN25917.1 KZS08995.1 AK417369 BAN20584.1 KK107488 QOIP01000012 EZA49951.1 RLU16080.1 GDHF01012739 JAI39575.1 GAKP01006623 JAC52329.1 GDHF01023682 GDHF01022729 JAI28632.1 JAI29585.1 AJVK01007913 AJVK01007914 KQ434954 KZC12527.1 CH933806 EDW16367.1 GL732610 EFX71388.1 GFTR01005350 JAW11076.1 GAMC01012792 GAMC01012790 JAB93763.1 GFXV01004761 MBW16566.1 KQ435794 KOX74119.1 PZQS01000009 PVD24420.1 KQ759866 OAD62493.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000283053
UP000007151
UP000075886
+ More
UP000007062 UP000075903 UP000000673 UP000076407 UP000075840 UP000075882 UP000075884 UP000075881 UP000075920 UP000002320 UP000075883 UP000075900 UP000076408 UP000008820 UP000030765 UP000075885 UP000075880 UP000245037 UP000242457 UP000069272 UP000078541 UP000078492 UP000005205 UP000027135 UP000075809 UP000078540 UP000008237 UP000002358 UP000092461 UP000235965 UP000198287 UP000075902 UP000094527 UP000030742 UP000095300 UP000000311 UP000078542 UP000095301 UP000000803 UP000192221 UP000001292 UP000007798 UP000007266 UP000002282 UP000053825 UP000015103 UP000008711 UP000076858 UP000053097 UP000279307 UP000092462 UP000076502 UP000009192 UP000000305 UP000053105 UP000245119
UP000007062 UP000075903 UP000000673 UP000076407 UP000075840 UP000075882 UP000075884 UP000075881 UP000075920 UP000002320 UP000075883 UP000075900 UP000076408 UP000008820 UP000030765 UP000075885 UP000075880 UP000245037 UP000242457 UP000069272 UP000078541 UP000078492 UP000005205 UP000027135 UP000075809 UP000078540 UP000008237 UP000002358 UP000092461 UP000235965 UP000198287 UP000075902 UP000094527 UP000030742 UP000095300 UP000000311 UP000078542 UP000095301 UP000000803 UP000192221 UP000001292 UP000007798 UP000007266 UP000002282 UP000053825 UP000015103 UP000008711 UP000076858 UP000053097 UP000279307 UP000092462 UP000076502 UP000009192 UP000000305 UP000053105 UP000245119
Interpro
Gene 3D
CDD
ProteinModelPortal
H9IZR5
A0A194R8S3
A0A2H1V0P1
S4PDP5
A0A194PX57
A0A3S2NM84
+ More
A0A212FA10 A0A182QQ36 A0A1S4H3X9 Q7QDU3 A0A336MXN1 A0A182UX13 A0A336MZK5 W5J7J0 A0A182XB08 A0A182HX81 A0A182LN80 A0A182NDJ7 A0A2M3Z685 A0A182K007 A0A182W3R4 A0A2M4AEX7 A0A2M4BSD4 B0XA87 A0A182LUG9 A0A182RBE4 A0A182YLY1 T1E2S0 Q17D51 A0A084VUL4 A0A182PEU7 A0A1S4F7M2 A0A1Q3FG45 A0A023ERL5 A0A182JDB8 A0A336MUR1 A0A1B6L2R2 A0A2P8XMD5 U5ESA2 V9IHF4 A0A2A3E2G8 A0A182FM08 Q1HPJ1 A0A1B6GEQ8 A0A1B6EG39 A0A195F310 A0A195DCA7 A0A158NE37 A0A067RJD6 A0A151WFB4 A0A195BBX2 A0A0A9Y541 E2C3B2 K7IXW5 A0A1B0EZK0 A0A1Y1LZ82 A0A2J7PL96 A0A1L8E0L6 A0A226DAT4 A0A182U880 A0A1D2N7U1 E9J7R1 U4TT33 A0A1I8Q9Y7 E2AI43 A0A1L8ECM4 A0A2Z5RE12 T1PC65 A0A1L8ED34 A0A151IGB1 A0A0V0GB25 A0A1I8MP09 Q8IGJ4 Q7K5K3 A0A1W4U638 A0A0K8TSX5 B4HZ58 B4NBF7 D6X401 A0A023FCH4 B4PQ74 A0A0L7QL62 T1IFK5 B3P5L5 A0A0N8E2A6 R4WDA4 A0A026W2Y6 A0A0K8VLY4 A0A034WCB5 A0A0K8UPM2 A0A1B0DP56 A0A154PL96 B4K754 E9HA87 A0A224XF23 W8B9P1 A0A2H8TQV1 A0A0M9A1W7 A0A2T7NTB3 A0A310SVU9
A0A212FA10 A0A182QQ36 A0A1S4H3X9 Q7QDU3 A0A336MXN1 A0A182UX13 A0A336MZK5 W5J7J0 A0A182XB08 A0A182HX81 A0A182LN80 A0A182NDJ7 A0A2M3Z685 A0A182K007 A0A182W3R4 A0A2M4AEX7 A0A2M4BSD4 B0XA87 A0A182LUG9 A0A182RBE4 A0A182YLY1 T1E2S0 Q17D51 A0A084VUL4 A0A182PEU7 A0A1S4F7M2 A0A1Q3FG45 A0A023ERL5 A0A182JDB8 A0A336MUR1 A0A1B6L2R2 A0A2P8XMD5 U5ESA2 V9IHF4 A0A2A3E2G8 A0A182FM08 Q1HPJ1 A0A1B6GEQ8 A0A1B6EG39 A0A195F310 A0A195DCA7 A0A158NE37 A0A067RJD6 A0A151WFB4 A0A195BBX2 A0A0A9Y541 E2C3B2 K7IXW5 A0A1B0EZK0 A0A1Y1LZ82 A0A2J7PL96 A0A1L8E0L6 A0A226DAT4 A0A182U880 A0A1D2N7U1 E9J7R1 U4TT33 A0A1I8Q9Y7 E2AI43 A0A1L8ECM4 A0A2Z5RE12 T1PC65 A0A1L8ED34 A0A151IGB1 A0A0V0GB25 A0A1I8MP09 Q8IGJ4 Q7K5K3 A0A1W4U638 A0A0K8TSX5 B4HZ58 B4NBF7 D6X401 A0A023FCH4 B4PQ74 A0A0L7QL62 T1IFK5 B3P5L5 A0A0N8E2A6 R4WDA4 A0A026W2Y6 A0A0K8VLY4 A0A034WCB5 A0A0K8UPM2 A0A1B0DP56 A0A154PL96 B4K754 E9HA87 A0A224XF23 W8B9P1 A0A2H8TQV1 A0A0M9A1W7 A0A2T7NTB3 A0A310SVU9
PDB
3EXI
E-value=1.32108e-128,
Score=1177
Ontologies
PATHWAY
00010
Glycolysis / Gluconeogenesis - Bombyx mori (domestic silkworm)
00020 Citrate cycle (TCA cycle) - Bombyx mori (domestic silkworm)
00620 Pyruvate metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
00020 Citrate cycle (TCA cycle) - Bombyx mori (domestic silkworm)
00620 Pyruvate metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
GO
PANTHER
Topology
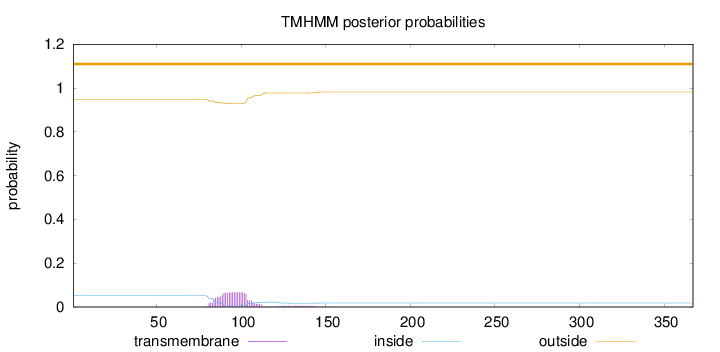
Length:
367
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.5572
Exp number, first 60 AAs:
0.00018
Total prob of N-in:
0.05191
outside
1 - 367
Population Genetic Test Statistics
Pi
4.080359
Theta
12.754612
Tajima's D
-2.00634
CLR
601.631141
CSRT
0.0124493775311234
Interpretation
Possibly Positive selection
