Pre Gene Modal
BGIBMGA014478
Annotation
PREDICTED:_saccharopine_dehydrogenase-like_oxidoreductase_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.856
Sequence
CDS
ATGCAGCTAAATTACAACGAACAAGCCGAAGACGCCGGCGTGTACGTCATCAGCGCCTGCGGATTTGACAGCATACCTTGTGACTTGGGCGTTATACACTTAATGAAAAACTTTGAGGGCGTGCTGAATTCTGTAGAGTCGTATATCATGATGAGGGTAGCGCCGGAGCACGCGGCGGAGGCGGCCCGCAGCGGCGTCCTGCACTACGGCACCTGGGAGTCCCTGGTGCACAGTCTGGCGAACAGTCACAAATTGAAGCCGCTGAGGGAACAGCTGTATCCCTCGCGGCTGCCCTCCTGCAAGCCCAAGCTCTGCCGACGATGGTACCACAGGAGGGGCGGCGCGTCGTGCCTGCCGTTCCTGGGCGCGGACGAGTCGGTGGTGCACCGCACGCAGCGCCACCTGCACGCGGCCGGCGCCCGCCCCGCGCAGCTCCGCGCCTACCTCGCGCTGCCCTCCGCACTGCACGCCGCGCTCCTCACGCTGGGCGCCCTCCTGCTGTACGTGCTGGCCCAGTGGAGCGTCACCAGGCGGCTGCTGCTCGACCACCCGCGGCTGTTCTCCGGCGGCATGGCGACCCGGCGCGGGCCCGCAGAGAACGTGATGCTCAACACCAAGTTCGAGGTGGAGCTGGTGGGGTCGGGCTGGGCCGCCGCGACCGACGCCGACGTCACCGAGCCGGACAAGAAGCTGGTTGTGAAGGTGTCGGGCTCCAACCCCGGGTACGGCGCCACGGTGACGGCGCTGCTGGTCTCCGCCCTCACTATACTGAACGAACAACACAAAATGCCTAAAAGCGGCGGGGTCCTGACAACCGGCGCCGCGTTCAAAGACACGAGCATCATTGATGTCCTCCACGAAAACGGCCTCAAGTTTGATATCAGTACGTAA
Protein
MQLNYNEQAEDAGVYVISACGFDSIPCDLGVIHLMKNFEGVLNSVESYIMMRVAPEHAAEAARSGVLHYGTWESLVHSLANSHKLKPLREQLYPSRLPSCKPKLCRRWYHRRGGASCLPFLGADESVVHRTQRHLHAAGARPAQLRAYLALPSALHAALLTLGALLLYVLAQWSVTRRLLLDHPRLFSGGMATRRGPAENVMLNTKFEVELVGSGWAAATDADVTEPDKKLVVKVSGSNPGYGATVTALLVSALTILNEQHKMPKSGGVLTTGAAFKDTSIIDVLHENGLKFDIST
Summary
Uniprot
H9JY60
A0A1E1WC13
A0A1E1VXL0
A0A2A4JQ28
A0A2H1WUH9
A0A212F873
+ More
A0A437BVQ6 S4PSD2 A0A194QFX5 A0A2A4JJC6 A0A437BM77 A0A1B6MG41 A0A182NRA6 A0A182VZZ4 U5EMX7 A0A182P3U7 Q17GW8 Q1HQU4 Q17GX0 A0A023ET52 A0A182Q1H0 A0A3B0JZV3 A0A182Y6P8 A0A023ERZ9 B0WA71 Q7QCJ0 A0A182TED9 A0A182XAJ3 A0A182HSD4 Q17GW9 A0A182RQK4 A0A182GXX7 A0A1S4F332 B4G2Z2 A0A2J7REF3 A0A2J7RED6 W8BYH8 B4K871 A0A182VKQ2 A0A182LEC5 B4NIZ0 A0A182GXX8 A0A1Q3FAZ6 A0A0L0CGL4 B3LVD9 A0A0K8U019 A0A084VTP8 A0A1B6H2S7 A0A023ERV2 Q297S7 B4JTQ0 A0A1B0AV82 B3P299 A0A034WNK5 A0A0M3QXV3 A0A0R3NKK4 A0A1Y9HDV2 A0A336M6G8 B4I3W8 A0A1W4UN15 A0A182M249 A0A182M578 A0A1A9VLX1 A0A182GXX9 A0A1B6HJK4 B4PV64 A0A182LCW1 Q7Q0G7 A0A182U5H1 A0A1A9ZQ48 A0A1B0CA41 A0A1W4UJW0 A0A182UWJ3 A0A1Q3FGN7 A0A1L8ECP5 A0A1L8ECR3 A0A084W724 A0A1L8ECP0 A0A182XKF8 A0A1A9W2U6 Q9VN86 A0A232EP89 K7ISA2 A0A1A9YFT7 B4QW83 A0A026W6W5 A0A2C9GQG8 B4NKS5 B4PNT0 A0A1J1J316 A0A3B0JDT7 B4LY94 B0WA72 W5JVK1 A0A1B0FM38 B4HGY8 A0A3F2Z105 A0A1W4WKW1 B3P1B4 Q9VG81
A0A437BVQ6 S4PSD2 A0A194QFX5 A0A2A4JJC6 A0A437BM77 A0A1B6MG41 A0A182NRA6 A0A182VZZ4 U5EMX7 A0A182P3U7 Q17GW8 Q1HQU4 Q17GX0 A0A023ET52 A0A182Q1H0 A0A3B0JZV3 A0A182Y6P8 A0A023ERZ9 B0WA71 Q7QCJ0 A0A182TED9 A0A182XAJ3 A0A182HSD4 Q17GW9 A0A182RQK4 A0A182GXX7 A0A1S4F332 B4G2Z2 A0A2J7REF3 A0A2J7RED6 W8BYH8 B4K871 A0A182VKQ2 A0A182LEC5 B4NIZ0 A0A182GXX8 A0A1Q3FAZ6 A0A0L0CGL4 B3LVD9 A0A0K8U019 A0A084VTP8 A0A1B6H2S7 A0A023ERV2 Q297S7 B4JTQ0 A0A1B0AV82 B3P299 A0A034WNK5 A0A0M3QXV3 A0A0R3NKK4 A0A1Y9HDV2 A0A336M6G8 B4I3W8 A0A1W4UN15 A0A182M249 A0A182M578 A0A1A9VLX1 A0A182GXX9 A0A1B6HJK4 B4PV64 A0A182LCW1 Q7Q0G7 A0A182U5H1 A0A1A9ZQ48 A0A1B0CA41 A0A1W4UJW0 A0A182UWJ3 A0A1Q3FGN7 A0A1L8ECP5 A0A1L8ECR3 A0A084W724 A0A1L8ECP0 A0A182XKF8 A0A1A9W2U6 Q9VN86 A0A232EP89 K7ISA2 A0A1A9YFT7 B4QW83 A0A026W6W5 A0A2C9GQG8 B4NKS5 B4PNT0 A0A1J1J316 A0A3B0JDT7 B4LY94 B0WA72 W5JVK1 A0A1B0FM38 B4HGY8 A0A3F2Z105 A0A1W4WKW1 B3P1B4 Q9VG81
Pubmed
19121390
22118469
23622113
26354079
17510324
17204158
+ More
24945155 25244985 12364791 14747013 17210077 26483478 17994087 24495485 18057021 20966253 26108605 24438588 15632085 25348373 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 28648823 20075255 24508170 30249741 20920257 23761445
24945155 25244985 12364791 14747013 17210077 26483478 17994087 24495485 18057021 20966253 26108605 24438588 15632085 25348373 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 28648823 20075255 24508170 30249741 20920257 23761445
EMBL
BABH01042326
GDQN01006555
JAT84499.1
GDQN01011625
JAT79429.1
NWSH01000840
+ More
PCG73919.1 ODYU01011153 SOQ56718.1 AGBW02009779 OWR49937.1 RSAL01000004 RVE54557.1 GAIX01013963 JAA78597.1 KQ459011 KPJ04387.1 NWSH01001373 PCG71492.1 RSAL01000033 RVE51461.1 GEBQ01005101 JAT34876.1 GANO01000821 JAB59050.1 CH477255 EAT45887.1 DQ440350 ABF18383.1 EAT45884.1 EAT45885.1 GAPW01001607 JAC11991.1 AXCN02001467 OUUW01000005 SPP81040.1 GAPW01001608 JAC11990.1 DS231869 EDS40900.1 AAAB01008859 EAA08164.4 EGK96812.1 APCN01000262 APCN01000263 EAT45886.1 JXUM01096451 KQ564265 KXJ72502.1 CH479179 EDW24187.1 NEVH01005277 PNF39200.1 PNF39201.1 GAMC01008199 JAB98356.1 CH933806 EDW15425.1 KRG01546.1 KRG01547.1 CH964272 EDW84892.1 JXUM01096456 KXJ72503.1 GFDL01010299 JAV24746.1 JRES01000409 KNC31558.1 CH902617 EDV41467.1 GDHF01032451 GDHF01014080 JAI19863.1 JAI38234.1 ATLV01016383 KE525080 KFB41342.1 GECZ01000812 JAS68957.1 GAPW01001651 JAC11947.1 CM000070 EAL28128.1 CH916374 EDV91479.1 JXJN01004067 CH954181 EDV47849.1 GAKP01003207 JAC55745.1 CP012526 ALC46573.1 KRS99812.1 UFQS01000622 UFQT01000622 SSX05497.1 SSX25856.1 CH480821 EDW54911.1 AXCM01004705 AXCM01000655 KXJ72504.1 GECU01032858 JAS74848.1 CM000160 EDW95733.1 AAAB01008982 EAA14649.4 AJWK01003261 GFDL01008325 JAV26720.1 GFDG01002289 JAV16510.1 GFDG01002288 JAV16511.1 ATLV01021094 KE525312 KFB46018.1 GFDG01002291 JAV16508.1 AE014297 AY075205 AAF52062.1 AAL68073.1 NNAY01002972 OXU20199.1 CM000364 EDX11682.1 KK107372 QOIP01000004 EZA51718.1 RLU23201.1 RLU23405.1 APCN01002169 EDW84136.1 EDW97095.1 CVRI01000066 CRL05862.1 SPP80487.1 CH940650 EDW67982.1 KRF83597.1 EDS40901.1 ADMH02000300 ETN67050.1 CCAG010005444 CH480815 EDW42459.1 EDV49373.1 BT024191 AAF54804.1 ABC86253.1
PCG73919.1 ODYU01011153 SOQ56718.1 AGBW02009779 OWR49937.1 RSAL01000004 RVE54557.1 GAIX01013963 JAA78597.1 KQ459011 KPJ04387.1 NWSH01001373 PCG71492.1 RSAL01000033 RVE51461.1 GEBQ01005101 JAT34876.1 GANO01000821 JAB59050.1 CH477255 EAT45887.1 DQ440350 ABF18383.1 EAT45884.1 EAT45885.1 GAPW01001607 JAC11991.1 AXCN02001467 OUUW01000005 SPP81040.1 GAPW01001608 JAC11990.1 DS231869 EDS40900.1 AAAB01008859 EAA08164.4 EGK96812.1 APCN01000262 APCN01000263 EAT45886.1 JXUM01096451 KQ564265 KXJ72502.1 CH479179 EDW24187.1 NEVH01005277 PNF39200.1 PNF39201.1 GAMC01008199 JAB98356.1 CH933806 EDW15425.1 KRG01546.1 KRG01547.1 CH964272 EDW84892.1 JXUM01096456 KXJ72503.1 GFDL01010299 JAV24746.1 JRES01000409 KNC31558.1 CH902617 EDV41467.1 GDHF01032451 GDHF01014080 JAI19863.1 JAI38234.1 ATLV01016383 KE525080 KFB41342.1 GECZ01000812 JAS68957.1 GAPW01001651 JAC11947.1 CM000070 EAL28128.1 CH916374 EDV91479.1 JXJN01004067 CH954181 EDV47849.1 GAKP01003207 JAC55745.1 CP012526 ALC46573.1 KRS99812.1 UFQS01000622 UFQT01000622 SSX05497.1 SSX25856.1 CH480821 EDW54911.1 AXCM01004705 AXCM01000655 KXJ72504.1 GECU01032858 JAS74848.1 CM000160 EDW95733.1 AAAB01008982 EAA14649.4 AJWK01003261 GFDL01008325 JAV26720.1 GFDG01002289 JAV16510.1 GFDG01002288 JAV16511.1 ATLV01021094 KE525312 KFB46018.1 GFDG01002291 JAV16508.1 AE014297 AY075205 AAF52062.1 AAL68073.1 NNAY01002972 OXU20199.1 CM000364 EDX11682.1 KK107372 QOIP01000004 EZA51718.1 RLU23201.1 RLU23405.1 APCN01002169 EDW84136.1 EDW97095.1 CVRI01000066 CRL05862.1 SPP80487.1 CH940650 EDW67982.1 KRF83597.1 EDS40901.1 ADMH02000300 ETN67050.1 CCAG010005444 CH480815 EDW42459.1 EDV49373.1 BT024191 AAF54804.1 ABC86253.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000283053
UP000053268
UP000075884
+ More
UP000075920 UP000075885 UP000008820 UP000075886 UP000268350 UP000076408 UP000002320 UP000007062 UP000075902 UP000076407 UP000075840 UP000075900 UP000069940 UP000249989 UP000008744 UP000235965 UP000009192 UP000075903 UP000075882 UP000007798 UP000037069 UP000007801 UP000030765 UP000001819 UP000001070 UP000092460 UP000008711 UP000092553 UP000001292 UP000192221 UP000075883 UP000078200 UP000002282 UP000092445 UP000092461 UP000091820 UP000000803 UP000215335 UP000002358 UP000092443 UP000000304 UP000053097 UP000279307 UP000183832 UP000008792 UP000000673 UP000092444 UP000192223
UP000075920 UP000075885 UP000008820 UP000075886 UP000268350 UP000076408 UP000002320 UP000007062 UP000075902 UP000076407 UP000075840 UP000075900 UP000069940 UP000249989 UP000008744 UP000235965 UP000009192 UP000075903 UP000075882 UP000007798 UP000037069 UP000007801 UP000030765 UP000001819 UP000001070 UP000092460 UP000008711 UP000092553 UP000001292 UP000192221 UP000075883 UP000078200 UP000002282 UP000092445 UP000092461 UP000091820 UP000000803 UP000215335 UP000002358 UP000092443 UP000000304 UP000053097 UP000279307 UP000183832 UP000008792 UP000000673 UP000092444 UP000192223
Interpro
ProteinModelPortal
H9JY60
A0A1E1WC13
A0A1E1VXL0
A0A2A4JQ28
A0A2H1WUH9
A0A212F873
+ More
A0A437BVQ6 S4PSD2 A0A194QFX5 A0A2A4JJC6 A0A437BM77 A0A1B6MG41 A0A182NRA6 A0A182VZZ4 U5EMX7 A0A182P3U7 Q17GW8 Q1HQU4 Q17GX0 A0A023ET52 A0A182Q1H0 A0A3B0JZV3 A0A182Y6P8 A0A023ERZ9 B0WA71 Q7QCJ0 A0A182TED9 A0A182XAJ3 A0A182HSD4 Q17GW9 A0A182RQK4 A0A182GXX7 A0A1S4F332 B4G2Z2 A0A2J7REF3 A0A2J7RED6 W8BYH8 B4K871 A0A182VKQ2 A0A182LEC5 B4NIZ0 A0A182GXX8 A0A1Q3FAZ6 A0A0L0CGL4 B3LVD9 A0A0K8U019 A0A084VTP8 A0A1B6H2S7 A0A023ERV2 Q297S7 B4JTQ0 A0A1B0AV82 B3P299 A0A034WNK5 A0A0M3QXV3 A0A0R3NKK4 A0A1Y9HDV2 A0A336M6G8 B4I3W8 A0A1W4UN15 A0A182M249 A0A182M578 A0A1A9VLX1 A0A182GXX9 A0A1B6HJK4 B4PV64 A0A182LCW1 Q7Q0G7 A0A182U5H1 A0A1A9ZQ48 A0A1B0CA41 A0A1W4UJW0 A0A182UWJ3 A0A1Q3FGN7 A0A1L8ECP5 A0A1L8ECR3 A0A084W724 A0A1L8ECP0 A0A182XKF8 A0A1A9W2U6 Q9VN86 A0A232EP89 K7ISA2 A0A1A9YFT7 B4QW83 A0A026W6W5 A0A2C9GQG8 B4NKS5 B4PNT0 A0A1J1J316 A0A3B0JDT7 B4LY94 B0WA72 W5JVK1 A0A1B0FM38 B4HGY8 A0A3F2Z105 A0A1W4WKW1 B3P1B4 Q9VG81
A0A437BVQ6 S4PSD2 A0A194QFX5 A0A2A4JJC6 A0A437BM77 A0A1B6MG41 A0A182NRA6 A0A182VZZ4 U5EMX7 A0A182P3U7 Q17GW8 Q1HQU4 Q17GX0 A0A023ET52 A0A182Q1H0 A0A3B0JZV3 A0A182Y6P8 A0A023ERZ9 B0WA71 Q7QCJ0 A0A182TED9 A0A182XAJ3 A0A182HSD4 Q17GW9 A0A182RQK4 A0A182GXX7 A0A1S4F332 B4G2Z2 A0A2J7REF3 A0A2J7RED6 W8BYH8 B4K871 A0A182VKQ2 A0A182LEC5 B4NIZ0 A0A182GXX8 A0A1Q3FAZ6 A0A0L0CGL4 B3LVD9 A0A0K8U019 A0A084VTP8 A0A1B6H2S7 A0A023ERV2 Q297S7 B4JTQ0 A0A1B0AV82 B3P299 A0A034WNK5 A0A0M3QXV3 A0A0R3NKK4 A0A1Y9HDV2 A0A336M6G8 B4I3W8 A0A1W4UN15 A0A182M249 A0A182M578 A0A1A9VLX1 A0A182GXX9 A0A1B6HJK4 B4PV64 A0A182LCW1 Q7Q0G7 A0A182U5H1 A0A1A9ZQ48 A0A1B0CA41 A0A1W4UJW0 A0A182UWJ3 A0A1Q3FGN7 A0A1L8ECP5 A0A1L8ECR3 A0A084W724 A0A1L8ECP0 A0A182XKF8 A0A1A9W2U6 Q9VN86 A0A232EP89 K7ISA2 A0A1A9YFT7 B4QW83 A0A026W6W5 A0A2C9GQG8 B4NKS5 B4PNT0 A0A1J1J316 A0A3B0JDT7 B4LY94 B0WA72 W5JVK1 A0A1B0FM38 B4HGY8 A0A3F2Z105 A0A1W4WKW1 B3P1B4 Q9VG81
Ontologies
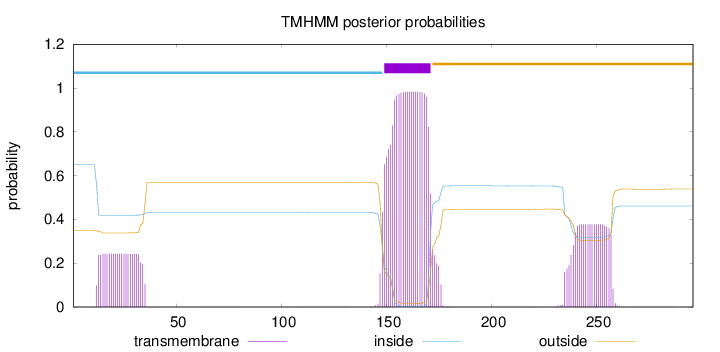
Topology
Length:
296
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
35.5323
Exp number, first 60 AAs:
5.43773
Total prob of N-in:
0.64948
inside
1 - 148
TMhelix
149 - 171
outside
172 - 296
Population Genetic Test Statistics
Pi
25.150203
Theta
24.108979
Tajima's D
0.189787
CLR
0.76121
CSRT
0.428628568571571
Interpretation
Uncertain
