Gene
KWMTBOMO03692
Annotation
PREDICTED:_saccharopine_dehydrogenase-like_oxidoreductase_[Bombyx_mori]
Full name
Lipid droplet localized protein
+ More
DNA helicase
DNA helicase
Location in the cell
Cytoplasmic Reliability : 1.06 Mitochondrial Reliability : 1.503
Sequence
CDS
ATGATTTTTGTACTATTCAAGGTTGGAGATAAGATGACCAGATTAGATCTGATAATATTCGGCGCGACCGGTTTCACCGGCAGCCGAGCGGTGCTGGAGATGATCAGGCTGTCCAAGAAGTATCCCGCTTTAACTTGGGGCATCGCTGGACGCTCCGAGAAAAAACTCGAAGAATTAAAACAAGAGGCTACTCAGAAAAGTGGTGAGGATCTTTAA
Protein
MIFVLFKVGDKMTRLDLIIFGATGFTGSRAVLEMIRLSKKYPALTWGIAGRSEKKLEELKQEATQKSGEDL
Summary
Catalytic Activity
ATP + H2O = ADP + H(+) + phosphate
Similarity
Belongs to the saccharopine dehydrogenase family.
Belongs to the RuvB family.
Belongs to the RuvB family.
Keywords
Alternative splicing
Complete proteome
Lipid droplet
Membrane
Reference proteome
Transmembrane
Transmembrane helix
Feature
chain Lipid droplet localized protein
splice variant In isoform b.
splice variant In isoform b.
Uniprot
A0A194QFX5
A0A194R9R0
A0A212F873
A0A2A4JJC6
A0A212EKN0
S4PSD2
+ More
A0A2A4JQ28 A0A437BM77 A0A437BVQ6 A0A1B6HWG5 A0A1B6IJI0 A0A182SN91 A0A1B6C8Z6 A0A1B6E0Q0 A0A1S4F332 Q17GW9 A0A2J7REE3 A0A0T6AVT0 A0A2J7REF3 A0A0K8TQ35 A0A2J7RED6 A0A1B6DUC4 A0A3F2Z105 B4KPY3 A0A2C9GQG8 A0A182UWJ3 A0A182U5H1 A0A182LCW1 Q7Q0G7 A0A182XKF8 A0A0T6AW50 A0A1J1J316 A0A182GXX8 G0MT30 A0A1B6HJK4 A0A182M249 G0PL87 A0A1B6MG41 A0A368G8U2 A0A1W4WKW1 E3LKM4 A0A2P4V2L3 A0A260ZWU8 A0A261BJ56 A0A1I7TYA1 A0A2G5TD76 A0A182J806 A0A1Y9HDV2 A8XRZ7 Q9GZE9 A0A067QRX6 A0A2P8ZJS7 W8BMR7 A0A1Y1N3F6 A0A0C2G6W3 Q17GW8 A0A0A1XFM1 A0A0L0C908 A0A2H2ILG5 A0A016TV51 V4B7C8 A0A2G8L7B1 A0A2H2HXA0 A0A2G8KGD3 A0A0D6LMM5 A0A1Y1MIX5 A0A1Y1MIU2 A0A1Y1MIK9 A0A182XXI8 A0A1Y1MMG4 A0A1Y1MLS2 A0A1Y1MIZ8 A0A158PIJ3 W2SNG8 B0WA72 A0A182M578 A0A1Y1MNW8 A0A0A1WM68 A0A0R3NKK4 U5EMX7 A0A1Y1NJ82 A0A182K3C8 A0A182PBG2 A0A3B0JDT7 A0A0N4XEX8 W5JVK1 A0A139WNZ6 A0A084VTP8 A0A182NL94 A0A182QET7 A0A034WNK5 B3M2Q1 A0A0D8YC32 A0A182GXX9 A0A182FAX4
A0A2A4JQ28 A0A437BM77 A0A437BVQ6 A0A1B6HWG5 A0A1B6IJI0 A0A182SN91 A0A1B6C8Z6 A0A1B6E0Q0 A0A1S4F332 Q17GW9 A0A2J7REE3 A0A0T6AVT0 A0A2J7REF3 A0A0K8TQ35 A0A2J7RED6 A0A1B6DUC4 A0A3F2Z105 B4KPY3 A0A2C9GQG8 A0A182UWJ3 A0A182U5H1 A0A182LCW1 Q7Q0G7 A0A182XKF8 A0A0T6AW50 A0A1J1J316 A0A182GXX8 G0MT30 A0A1B6HJK4 A0A182M249 G0PL87 A0A1B6MG41 A0A368G8U2 A0A1W4WKW1 E3LKM4 A0A2P4V2L3 A0A260ZWU8 A0A261BJ56 A0A1I7TYA1 A0A2G5TD76 A0A182J806 A0A1Y9HDV2 A8XRZ7 Q9GZE9 A0A067QRX6 A0A2P8ZJS7 W8BMR7 A0A1Y1N3F6 A0A0C2G6W3 Q17GW8 A0A0A1XFM1 A0A0L0C908 A0A2H2ILG5 A0A016TV51 V4B7C8 A0A2G8L7B1 A0A2H2HXA0 A0A2G8KGD3 A0A0D6LMM5 A0A1Y1MIX5 A0A1Y1MIU2 A0A1Y1MIK9 A0A182XXI8 A0A1Y1MMG4 A0A1Y1MLS2 A0A1Y1MIZ8 A0A158PIJ3 W2SNG8 B0WA72 A0A182M578 A0A1Y1MNW8 A0A0A1WM68 A0A0R3NKK4 U5EMX7 A0A1Y1NJ82 A0A182K3C8 A0A182PBG2 A0A3B0JDT7 A0A0N4XEX8 W5JVK1 A0A139WNZ6 A0A084VTP8 A0A182NL94 A0A182QET7 A0A034WNK5 B3M2Q1 A0A0D8YC32 A0A182GXX9 A0A182FAX4
EC Number
3.6.4.12
Pubmed
26354079
22118469
23622113
17510324
26369729
17994087
+ More
20966253 12364791 14747013 17210077 26483478 26114425 26394399 14624247 9851916 26121959 26025681 24845553 29403074 24495485 28004739 25830018 26108605 25730766 23254933 29023486 25244985 15632085 20920257 23761445 18362917 19820115 24438588 25348373 26856411
20966253 12364791 14747013 17210077 26483478 26114425 26394399 14624247 9851916 26121959 26025681 24845553 29403074 24495485 28004739 25830018 26108605 25730766 23254933 29023486 25244985 15632085 20920257 23761445 18362917 19820115 24438588 25348373 26856411
EMBL
KQ459011
KPJ04387.1
KQ460500
KPJ14244.1
AGBW02009779
OWR49937.1
+ More
NWSH01001373 PCG71492.1 AGBW02014259 OWR42008.1 GAIX01013963 JAA78597.1 NWSH01000840 PCG73919.1 RSAL01000033 RVE51461.1 RSAL01000004 RVE54557.1 GECU01028682 JAS79024.1 GECU01020630 JAS87076.1 GEDC01027499 GEDC01013409 JAS09799.1 JAS23889.1 GEDC01005792 JAS31506.1 CH477255 EAT45886.1 NEVH01005277 PNF39199.1 LJIG01022680 KRT79283.1 PNF39200.1 GDAI01001124 JAI16479.1 PNF39201.1 GEDC01022313 GEDC01008021 JAS14985.1 JAS29277.1 CH933808 EDW10260.1 APCN01002169 AAAB01008982 EAA14649.4 KRT79281.1 CVRI01000066 CRL05862.1 JXUM01096456 KQ564265 KXJ72503.1 GL379811 EGT43549.1 GECU01032858 JAS74848.1 AXCM01004705 GL381031 EGT34189.1 GEBQ01005101 JAT34876.1 JOJR01000338 RCN39425.1 DS268410 EFO99888.1 LFJK02001396 POM31533.1 NMWX01000033 OZF90082.1 NIPN01000125 OZG10361.1 PDUG01000005 PIC24996.1 HE601413 CAP35416.1 BX284605 KK853004 KDR12595.1 PYGN01000035 PSN56748.1 GAMC01008262 JAB98293.1 GEZM01014976 JAV91948.1 KN738734 KIH54654.1 EAT45887.1 GBXI01004535 JAD09757.1 JRES01000752 KNC28766.1 JARK01001411 EYC06616.1 KB203505 ESO84484.1 MRZV01000187 PIK56134.1 MRZV01000607 PIK47030.1 KE125047 EPB72448.1 GEZM01030324 JAV85631.1 GEZM01030321 GEZM01030318 JAV85634.1 GEZM01030326 JAV85629.1 GEZM01030320 JAV85635.1 GEZM01030325 GEZM01030317 JAV85630.1 GEZM01030323 GEZM01030322 JAV85633.1 UYYA01004060 VDM59218.1 KI668967 ETN70386.1 DS231869 EDS40901.1 AXCM01000655 GEZM01030319 JAV85636.1 GBXI01014148 JAD00144.1 CM000070 KRS99812.1 GANO01000821 JAB59050.1 GEZM01001299 JAV97972.1 OUUW01000005 SPP80487.1 UYSL01000726 VDL64314.1 ADMH02000300 ETN67050.1 KQ971311 KYB29495.1 ATLV01016383 KE525080 KFB41342.1 AXCN02000112 GAKP01003207 JAC55745.1 CH902617 EDV42372.1 KN716150 KJH53614.1 KXJ72504.1
NWSH01001373 PCG71492.1 AGBW02014259 OWR42008.1 GAIX01013963 JAA78597.1 NWSH01000840 PCG73919.1 RSAL01000033 RVE51461.1 RSAL01000004 RVE54557.1 GECU01028682 JAS79024.1 GECU01020630 JAS87076.1 GEDC01027499 GEDC01013409 JAS09799.1 JAS23889.1 GEDC01005792 JAS31506.1 CH477255 EAT45886.1 NEVH01005277 PNF39199.1 LJIG01022680 KRT79283.1 PNF39200.1 GDAI01001124 JAI16479.1 PNF39201.1 GEDC01022313 GEDC01008021 JAS14985.1 JAS29277.1 CH933808 EDW10260.1 APCN01002169 AAAB01008982 EAA14649.4 KRT79281.1 CVRI01000066 CRL05862.1 JXUM01096456 KQ564265 KXJ72503.1 GL379811 EGT43549.1 GECU01032858 JAS74848.1 AXCM01004705 GL381031 EGT34189.1 GEBQ01005101 JAT34876.1 JOJR01000338 RCN39425.1 DS268410 EFO99888.1 LFJK02001396 POM31533.1 NMWX01000033 OZF90082.1 NIPN01000125 OZG10361.1 PDUG01000005 PIC24996.1 HE601413 CAP35416.1 BX284605 KK853004 KDR12595.1 PYGN01000035 PSN56748.1 GAMC01008262 JAB98293.1 GEZM01014976 JAV91948.1 KN738734 KIH54654.1 EAT45887.1 GBXI01004535 JAD09757.1 JRES01000752 KNC28766.1 JARK01001411 EYC06616.1 KB203505 ESO84484.1 MRZV01000187 PIK56134.1 MRZV01000607 PIK47030.1 KE125047 EPB72448.1 GEZM01030324 JAV85631.1 GEZM01030321 GEZM01030318 JAV85634.1 GEZM01030326 JAV85629.1 GEZM01030320 JAV85635.1 GEZM01030325 GEZM01030317 JAV85630.1 GEZM01030323 GEZM01030322 JAV85633.1 UYYA01004060 VDM59218.1 KI668967 ETN70386.1 DS231869 EDS40901.1 AXCM01000655 GEZM01030319 JAV85636.1 GBXI01014148 JAD00144.1 CM000070 KRS99812.1 GANO01000821 JAB59050.1 GEZM01001299 JAV97972.1 OUUW01000005 SPP80487.1 UYSL01000726 VDL64314.1 ADMH02000300 ETN67050.1 KQ971311 KYB29495.1 ATLV01016383 KE525080 KFB41342.1 AXCN02000112 GAKP01003207 JAC55745.1 CH902617 EDV42372.1 KN716150 KJH53614.1 KXJ72504.1
Proteomes
UP000053268
UP000053240
UP000007151
UP000218220
UP000283053
UP000075901
+ More
UP000008820 UP000235965 UP000075920 UP000009192 UP000075840 UP000075903 UP000075902 UP000075882 UP000007062 UP000076407 UP000183832 UP000069940 UP000249989 UP000008068 UP000075883 UP000252519 UP000192223 UP000008281 UP000237256 UP000216624 UP000216463 UP000095282 UP000230233 UP000075880 UP000075900 UP000008549 UP000001940 UP000027135 UP000245037 UP000037069 UP000005237 UP000024635 UP000030746 UP000230750 UP000076408 UP000050601 UP000267027 UP000002320 UP000001819 UP000075881 UP000075885 UP000268350 UP000038043 UP000271162 UP000000673 UP000007266 UP000030765 UP000075884 UP000075886 UP000007801 UP000053766 UP000069272
UP000008820 UP000235965 UP000075920 UP000009192 UP000075840 UP000075903 UP000075902 UP000075882 UP000007062 UP000076407 UP000183832 UP000069940 UP000249989 UP000008068 UP000075883 UP000252519 UP000192223 UP000008281 UP000237256 UP000216624 UP000216463 UP000095282 UP000230233 UP000075880 UP000075900 UP000008549 UP000001940 UP000027135 UP000245037 UP000037069 UP000005237 UP000024635 UP000030746 UP000230750 UP000076408 UP000050601 UP000267027 UP000002320 UP000001819 UP000075881 UP000075885 UP000268350 UP000038043 UP000271162 UP000000673 UP000007266 UP000030765 UP000075884 UP000075886 UP000007801 UP000053766 UP000069272
PRIDE
Interpro
ProteinModelPortal
A0A194QFX5
A0A194R9R0
A0A212F873
A0A2A4JJC6
A0A212EKN0
S4PSD2
+ More
A0A2A4JQ28 A0A437BM77 A0A437BVQ6 A0A1B6HWG5 A0A1B6IJI0 A0A182SN91 A0A1B6C8Z6 A0A1B6E0Q0 A0A1S4F332 Q17GW9 A0A2J7REE3 A0A0T6AVT0 A0A2J7REF3 A0A0K8TQ35 A0A2J7RED6 A0A1B6DUC4 A0A3F2Z105 B4KPY3 A0A2C9GQG8 A0A182UWJ3 A0A182U5H1 A0A182LCW1 Q7Q0G7 A0A182XKF8 A0A0T6AW50 A0A1J1J316 A0A182GXX8 G0MT30 A0A1B6HJK4 A0A182M249 G0PL87 A0A1B6MG41 A0A368G8U2 A0A1W4WKW1 E3LKM4 A0A2P4V2L3 A0A260ZWU8 A0A261BJ56 A0A1I7TYA1 A0A2G5TD76 A0A182J806 A0A1Y9HDV2 A8XRZ7 Q9GZE9 A0A067QRX6 A0A2P8ZJS7 W8BMR7 A0A1Y1N3F6 A0A0C2G6W3 Q17GW8 A0A0A1XFM1 A0A0L0C908 A0A2H2ILG5 A0A016TV51 V4B7C8 A0A2G8L7B1 A0A2H2HXA0 A0A2G8KGD3 A0A0D6LMM5 A0A1Y1MIX5 A0A1Y1MIU2 A0A1Y1MIK9 A0A182XXI8 A0A1Y1MMG4 A0A1Y1MLS2 A0A1Y1MIZ8 A0A158PIJ3 W2SNG8 B0WA72 A0A182M578 A0A1Y1MNW8 A0A0A1WM68 A0A0R3NKK4 U5EMX7 A0A1Y1NJ82 A0A182K3C8 A0A182PBG2 A0A3B0JDT7 A0A0N4XEX8 W5JVK1 A0A139WNZ6 A0A084VTP8 A0A182NL94 A0A182QET7 A0A034WNK5 B3M2Q1 A0A0D8YC32 A0A182GXX9 A0A182FAX4
A0A2A4JQ28 A0A437BM77 A0A437BVQ6 A0A1B6HWG5 A0A1B6IJI0 A0A182SN91 A0A1B6C8Z6 A0A1B6E0Q0 A0A1S4F332 Q17GW9 A0A2J7REE3 A0A0T6AVT0 A0A2J7REF3 A0A0K8TQ35 A0A2J7RED6 A0A1B6DUC4 A0A3F2Z105 B4KPY3 A0A2C9GQG8 A0A182UWJ3 A0A182U5H1 A0A182LCW1 Q7Q0G7 A0A182XKF8 A0A0T6AW50 A0A1J1J316 A0A182GXX8 G0MT30 A0A1B6HJK4 A0A182M249 G0PL87 A0A1B6MG41 A0A368G8U2 A0A1W4WKW1 E3LKM4 A0A2P4V2L3 A0A260ZWU8 A0A261BJ56 A0A1I7TYA1 A0A2G5TD76 A0A182J806 A0A1Y9HDV2 A8XRZ7 Q9GZE9 A0A067QRX6 A0A2P8ZJS7 W8BMR7 A0A1Y1N3F6 A0A0C2G6W3 Q17GW8 A0A0A1XFM1 A0A0L0C908 A0A2H2ILG5 A0A016TV51 V4B7C8 A0A2G8L7B1 A0A2H2HXA0 A0A2G8KGD3 A0A0D6LMM5 A0A1Y1MIX5 A0A1Y1MIU2 A0A1Y1MIK9 A0A182XXI8 A0A1Y1MMG4 A0A1Y1MLS2 A0A1Y1MIZ8 A0A158PIJ3 W2SNG8 B0WA72 A0A182M578 A0A1Y1MNW8 A0A0A1WM68 A0A0R3NKK4 U5EMX7 A0A1Y1NJ82 A0A182K3C8 A0A182PBG2 A0A3B0JDT7 A0A0N4XEX8 W5JVK1 A0A139WNZ6 A0A084VTP8 A0A182NL94 A0A182QET7 A0A034WNK5 B3M2Q1 A0A0D8YC32 A0A182GXX9 A0A182FAX4
Ontologies
GO
PANTHER
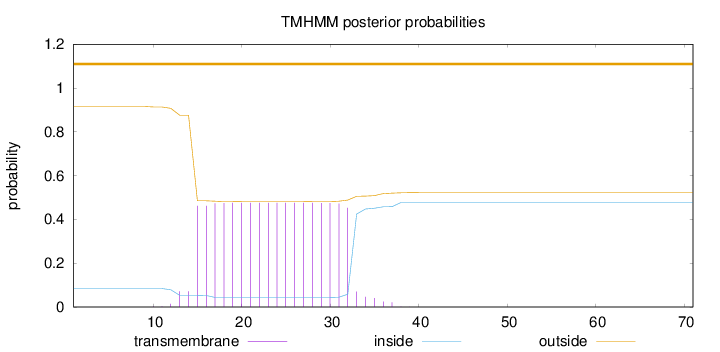
Topology
Subcellular location
Membrane
Lipid droplet
Nucleus
Lipid droplet
Nucleus
Length:
71
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
8.86659
Exp number, first 60 AAs:
8.86659
Total prob of N-in:
0.08426
outside
1 - 71
Population Genetic Test Statistics
Pi
22.961628
Theta
19.64192
Tajima's D
-1.667706
CLR
2.50975
CSRT
0.0413479326033698
Interpretation
Uncertain
