Gene
KWMTBOMO03690
Pre Gene Modal
BGIBMGA014352
Annotation
PREDICTED:_glutamate_dehydrogenase?_mitochondrial-like_[Papilio_machaon]
Full name
Glutamate dehydrogenase
+ More
Glutamate dehydrogenase 2, mitochondrial
Glutamate dehydrogenase, mitochondrial
Glutamate dehydrogenase 1, mitochondrial
Glutamate dehydrogenase 2, mitochondrial
Glutamate dehydrogenase, mitochondrial
Glutamate dehydrogenase 1, mitochondrial
Location in the cell
Mitochondrial Reliability : 2.084
Sequence
CDS
ATGAATATAGGTGTAGCGATAAATCCAAAACAATATACTATCGCTGAACTGCAGAGAATAACGAGAAGATATACTTTAGAACTTGCGAAGAAGAATTATATCGGTGCCGGCATCGACGTTCCAGCACCCGACGTGAACACGTCCGGTCGGGAGATGTCCTGGATCGTGGACACCTACATCAAGACGCTTGGTAAACTGTCACCATCGCTTCACCCAGCAAGAGCTACAAAGCCCTCGAGTTCCTTACAATTATTATTCATCGGCCATCAAATAACTTATATCATCTATCAATGTAGCCAATAA
Protein
MNIGVAINPKQYTIAELQRITRRYTLELAKKNYIGAGIDVPAPDVNTSGREMSWIVDTYIKTLGKLSPSLHPARATKPSSSLQLLFIGHQITYIIYQCSQ
Summary
Description
Mitochondrial glutamate dehydrogenase that converts L-glutamate into alpha-ketoglutarate. Plays a key role in glutamine anaplerosis by producing alpha-ketoglutarate, an important intermediate in the tricarboxylic acid cycle. May be involved in learning and memory reactions by increasing the turnover of the excitatory neurotransmitter glutamate (By similarity).
Catalytic Activity
H2O + L-glutamate + NAD(+) = 2-oxoglutarate + H(+) + NADH + NH4(+)
H2O + L-glutamate + NADP(+) = 2-oxoglutarate + H(+) + NADPH + NH4(+)
H2O + L-glutamate + NADP(+) = 2-oxoglutarate + H(+) + NADPH + NH4(+)
Subunit
Homohexamer.
Miscellaneous
ADP can occupy the NADH binding site and activate the enzyme.
Similarity
Belongs to the Glu/Leu/Phe/Val dehydrogenases family.
Keywords
ADP-ribosylation
Mitochondrion
NADP
Oxidoreductase
Transit peptide
Alternative splicing
ATP-binding
Complete proteome
GTP-binding
NAD
Nucleotide-binding
Reference proteome
3D-structure
Acetylation
Direct protein sequencing
Disease mutation
Phosphoprotein
Feature
chain Glutamate dehydrogenase 2, mitochondrial
splice variant In isoform C.
sequence variant In HHF6; diminished sensitivity to GTP.
splice variant In isoform C.
sequence variant In HHF6; diminished sensitivity to GTP.
Uniprot
A0A1B6I3X7
A0A1B6MCH4
A0A0A0N1J8
A0A1B6H6A7
A0A1B6LL06
A0A1B0G229
+ More
A0A0J7KNC5 A0A226DZK2 A0A1B6GVV3 A0A1B6EYJ5 I0IVB6 A0A1A9W5Q9 A0A2R7W1V0 A0A2H1V7P9 A0A0L7RDB5 A0A2P8ZB91 A0A3B0JPB7 A0A0U2U7H1 A0A0C4DHE7 R9QZD7 A0A0U2ICK1 A0A3P4NIB8 A0A0U2USC9 A0A0U2SKB2 A0A1A9VDQ7 A0A0U2SMK1 A0A0U2U3U9 A0A0U2UL37 D3TQR2 A0A1A9ZSU9 A0A232F5M0 K7IYA3 A0A1A9Y0I5 A0A1B0B2Z8 A0A0U2SWY2 A0A3B0JK98 Q8IMY1 A0A0K8UDM9 A0A1L8EA44 A0A3Q0E467 A0A171A2I5 A0A1L8EAH7 A0A2J7PVK0 H2PWN9 Q64I00 G1S9P7 Q64HZ9 A0A1I9WLI4 M1ERX9 A0A0K8SKG2 A0A1B6CCS9 A0A2S2PHU9 T1PBD9 A0A091KT94 Q59FQ4 B3M317 A0A1I8NSV1 A0A1L8EEM7 V9ICN1 A0A3B0JQU1 P54385-2 A0A1W4UJV6 I5ANX6 B4QSD4 Q298M7 B4G4C1 A0A3B0KCM3 T1P987 B4PL48 B3P8B7 E2C3C7 B4N9E8 B4HG10 A0A1S4FQT2 P54385 B4M0C5 A0A2U3YY02 A0A3Q0E0N0 A0A1L8EER7 A0A0Q9WUT7 A0A1I8NSU8 A0A2S2PUP3 P00367-3 B4K740 T1HLZ6
A0A0J7KNC5 A0A226DZK2 A0A1B6GVV3 A0A1B6EYJ5 I0IVB6 A0A1A9W5Q9 A0A2R7W1V0 A0A2H1V7P9 A0A0L7RDB5 A0A2P8ZB91 A0A3B0JPB7 A0A0U2U7H1 A0A0C4DHE7 R9QZD7 A0A0U2ICK1 A0A3P4NIB8 A0A0U2USC9 A0A0U2SKB2 A0A1A9VDQ7 A0A0U2SMK1 A0A0U2U3U9 A0A0U2UL37 D3TQR2 A0A1A9ZSU9 A0A232F5M0 K7IYA3 A0A1A9Y0I5 A0A1B0B2Z8 A0A0U2SWY2 A0A3B0JK98 Q8IMY1 A0A0K8UDM9 A0A1L8EA44 A0A3Q0E467 A0A171A2I5 A0A1L8EAH7 A0A2J7PVK0 H2PWN9 Q64I00 G1S9P7 Q64HZ9 A0A1I9WLI4 M1ERX9 A0A0K8SKG2 A0A1B6CCS9 A0A2S2PHU9 T1PBD9 A0A091KT94 Q59FQ4 B3M317 A0A1I8NSV1 A0A1L8EEM7 V9ICN1 A0A3B0JQU1 P54385-2 A0A1W4UJV6 I5ANX6 B4QSD4 Q298M7 B4G4C1 A0A3B0KCM3 T1P987 B4PL48 B3P8B7 E2C3C7 B4N9E8 B4HG10 A0A1S4FQT2 P54385 B4M0C5 A0A2U3YY02 A0A3Q0E0N0 A0A1L8EER7 A0A0Q9WUT7 A0A1I8NSU8 A0A2S2PUP3 P00367-3 B4K740 T1HLZ6
EC Number
1.4.1.3
Pubmed
29403074
10731132
12537568
12537572
12537573
12537574
+ More
16110336 17569856 17569867 24330026 20353571 28648823 20075255 26109357 26109356 15378063 27538518 23236062 26823975 25315136 17994087 10992165 12537569 6810872 15632085 17550304 20798317 17510324 3426581 3377777 3399399 3368458 8486350 14702039 15164054 15489334 429360 1286669 3585334 8314555 16023112 19608861 21269460 24275569 25944712 11254391 11903050 16959573 12054821 12653548 10338089 9571255 10636977 11214910 11297618
16110336 17569856 17569867 24330026 20353571 28648823 20075255 26109357 26109356 15378063 27538518 23236062 26823975 25315136 17994087 10992165 12537569 6810872 15632085 17550304 20798317 17510324 3426581 3377777 3399399 3368458 8486350 14702039 15164054 15489334 429360 1286669 3585334 8314555 16023112 19608861 21269460 24275569 25944712 11254391 11903050 16959573 12054821 12653548 10338089 9571255 10636977 11214910 11297618
EMBL
GECU01026111
JAS81595.1
GEBQ01006359
JAT33618.1
JQ861712
AGC92240.1
+ More
GECU01037447 JAS70259.1 GEBQ01015586 JAT24391.1 CCAG010010758 LBMM01005184 KMQ91741.1 LNIX01000008 OXA50902.1 GECZ01003209 GECZ01000641 JAS66560.1 JAS69128.1 GECZ01026752 GECZ01010743 JAS43017.1 JAS59026.1 AB706394 BAM09215.1 KK854250 PTY13716.1 ODYU01001114 SOQ36873.1 KQ414615 KOC68746.1 PYGN01000117 PSN53762.1 OUUW01000007 SPP82763.1 KU234365 ALR85856.1 AE014297 AAN13963.3 JX125591 AGG09855.1 KU234369 ALR85860.1 CYRY02020334 VCW97027.1 KU234368 ALR85859.1 KU234367 ALR85858.1 KU234366 ALR85857.1 KU234364 ALR85855.1 KU234363 ALR85854.1 EZ423764 ADD20040.1 NNAY01000875 OXU26104.1 JXJN01007741 JXJN01007742 JXJN01007743 KU234370 ALR85861.1 SPP82764.1 BT128894 AAN13962.1 AEO46471.1 GDHF01027829 JAI24485.1 GFDG01003225 JAV15574.1 GEMB01001566 JAS01593.1 GFDG01003226 JAV15573.1 NEVH01020956 PNF20356.1 ABGA01245569 AY588268 ADFV01008835 AY588269 KU932368 APA34004.1 JP010539 AER99136.1 GBRD01012534 GDHC01009264 JAG53290.1 JAQ09365.1 GEDC01026153 JAS11145.1 GGMR01012767 GGMR01016376 MBY25386.1 MBY28995.1 KA645293 AFP59922.1 KK753902 KFP42693.1 AB209406 BAD92643.1 CH902617 EDV42417.1 GFDG01001617 JAV17182.1 JR038794 AEY58417.1 SPP82762.1 Y11314 Z29062 AY061323 BT001501 CM000070 EIM52661.1 CM000364 EDX14133.1 EAL27928.2 CH479179 EDW24469.1 SPP82761.1 KA645292 AFP59921.1 CM000160 EDW99033.1 CH954182 EDV53941.1 GL452320 EFN77465.1 CH964232 EDW80581.1 CH480815 EDW43403.1 CH940650 EDW67287.1 GFDG01001619 JAV17180.1 KRF99304.1 GGMS01000045 MBY69248.1 X07674 M20867 M37154 X07769 J03248 X66300 X66301 X66302 X66303 X66304 X66305 X66306 X66307 X66308 X66309 X66311 X66312 AK122685 AK294685 AL136982 CH471142 BC040132 BC112946 X67491 CH933806 EDW16353.1 ACPB03009469 ACPB03009470 ACPB03009471 ACPB03009472
GECU01037447 JAS70259.1 GEBQ01015586 JAT24391.1 CCAG010010758 LBMM01005184 KMQ91741.1 LNIX01000008 OXA50902.1 GECZ01003209 GECZ01000641 JAS66560.1 JAS69128.1 GECZ01026752 GECZ01010743 JAS43017.1 JAS59026.1 AB706394 BAM09215.1 KK854250 PTY13716.1 ODYU01001114 SOQ36873.1 KQ414615 KOC68746.1 PYGN01000117 PSN53762.1 OUUW01000007 SPP82763.1 KU234365 ALR85856.1 AE014297 AAN13963.3 JX125591 AGG09855.1 KU234369 ALR85860.1 CYRY02020334 VCW97027.1 KU234368 ALR85859.1 KU234367 ALR85858.1 KU234366 ALR85857.1 KU234364 ALR85855.1 KU234363 ALR85854.1 EZ423764 ADD20040.1 NNAY01000875 OXU26104.1 JXJN01007741 JXJN01007742 JXJN01007743 KU234370 ALR85861.1 SPP82764.1 BT128894 AAN13962.1 AEO46471.1 GDHF01027829 JAI24485.1 GFDG01003225 JAV15574.1 GEMB01001566 JAS01593.1 GFDG01003226 JAV15573.1 NEVH01020956 PNF20356.1 ABGA01245569 AY588268 ADFV01008835 AY588269 KU932368 APA34004.1 JP010539 AER99136.1 GBRD01012534 GDHC01009264 JAG53290.1 JAQ09365.1 GEDC01026153 JAS11145.1 GGMR01012767 GGMR01016376 MBY25386.1 MBY28995.1 KA645293 AFP59922.1 KK753902 KFP42693.1 AB209406 BAD92643.1 CH902617 EDV42417.1 GFDG01001617 JAV17182.1 JR038794 AEY58417.1 SPP82762.1 Y11314 Z29062 AY061323 BT001501 CM000070 EIM52661.1 CM000364 EDX14133.1 EAL27928.2 CH479179 EDW24469.1 SPP82761.1 KA645292 AFP59921.1 CM000160 EDW99033.1 CH954182 EDV53941.1 GL452320 EFN77465.1 CH964232 EDW80581.1 CH480815 EDW43403.1 CH940650 EDW67287.1 GFDG01001619 JAV17180.1 KRF99304.1 GGMS01000045 MBY69248.1 X07674 M20867 M37154 X07769 J03248 X66300 X66301 X66302 X66303 X66304 X66305 X66306 X66307 X66308 X66309 X66311 X66312 AK122685 AK294685 AL136982 CH471142 BC040132 BC112946 X67491 CH933806 EDW16353.1 ACPB03009469 ACPB03009470 ACPB03009471 ACPB03009472
Proteomes
UP000092444
UP000036403
UP000198287
UP000091820
UP000053825
UP000245037
+ More
UP000268350 UP000000803 UP000078200 UP000092445 UP000215335 UP000002358 UP000092443 UP000092460 UP000189704 UP000235965 UP000001595 UP000001073 UP000095301 UP000007801 UP000095300 UP000192221 UP000001819 UP000000304 UP000008744 UP000002282 UP000008711 UP000008237 UP000007798 UP000001292 UP000008792 UP000245341 UP000005640 UP000009192 UP000015103
UP000268350 UP000000803 UP000078200 UP000092445 UP000215335 UP000002358 UP000092443 UP000092460 UP000189704 UP000235965 UP000001595 UP000001073 UP000095301 UP000007801 UP000095300 UP000192221 UP000001819 UP000000304 UP000008744 UP000002282 UP000008711 UP000008237 UP000007798 UP000001292 UP000008792 UP000245341 UP000005640 UP000009192 UP000015103
Interpro
SUPFAM
SSF51735
SSF51735
ProteinModelPortal
A0A1B6I3X7
A0A1B6MCH4
A0A0A0N1J8
A0A1B6H6A7
A0A1B6LL06
A0A1B0G229
+ More
A0A0J7KNC5 A0A226DZK2 A0A1B6GVV3 A0A1B6EYJ5 I0IVB6 A0A1A9W5Q9 A0A2R7W1V0 A0A2H1V7P9 A0A0L7RDB5 A0A2P8ZB91 A0A3B0JPB7 A0A0U2U7H1 A0A0C4DHE7 R9QZD7 A0A0U2ICK1 A0A3P4NIB8 A0A0U2USC9 A0A0U2SKB2 A0A1A9VDQ7 A0A0U2SMK1 A0A0U2U3U9 A0A0U2UL37 D3TQR2 A0A1A9ZSU9 A0A232F5M0 K7IYA3 A0A1A9Y0I5 A0A1B0B2Z8 A0A0U2SWY2 A0A3B0JK98 Q8IMY1 A0A0K8UDM9 A0A1L8EA44 A0A3Q0E467 A0A171A2I5 A0A1L8EAH7 A0A2J7PVK0 H2PWN9 Q64I00 G1S9P7 Q64HZ9 A0A1I9WLI4 M1ERX9 A0A0K8SKG2 A0A1B6CCS9 A0A2S2PHU9 T1PBD9 A0A091KT94 Q59FQ4 B3M317 A0A1I8NSV1 A0A1L8EEM7 V9ICN1 A0A3B0JQU1 P54385-2 A0A1W4UJV6 I5ANX6 B4QSD4 Q298M7 B4G4C1 A0A3B0KCM3 T1P987 B4PL48 B3P8B7 E2C3C7 B4N9E8 B4HG10 A0A1S4FQT2 P54385 B4M0C5 A0A2U3YY02 A0A3Q0E0N0 A0A1L8EER7 A0A0Q9WUT7 A0A1I8NSU8 A0A2S2PUP3 P00367-3 B4K740 T1HLZ6
A0A0J7KNC5 A0A226DZK2 A0A1B6GVV3 A0A1B6EYJ5 I0IVB6 A0A1A9W5Q9 A0A2R7W1V0 A0A2H1V7P9 A0A0L7RDB5 A0A2P8ZB91 A0A3B0JPB7 A0A0U2U7H1 A0A0C4DHE7 R9QZD7 A0A0U2ICK1 A0A3P4NIB8 A0A0U2USC9 A0A0U2SKB2 A0A1A9VDQ7 A0A0U2SMK1 A0A0U2U3U9 A0A0U2UL37 D3TQR2 A0A1A9ZSU9 A0A232F5M0 K7IYA3 A0A1A9Y0I5 A0A1B0B2Z8 A0A0U2SWY2 A0A3B0JK98 Q8IMY1 A0A0K8UDM9 A0A1L8EA44 A0A3Q0E467 A0A171A2I5 A0A1L8EAH7 A0A2J7PVK0 H2PWN9 Q64I00 G1S9P7 Q64HZ9 A0A1I9WLI4 M1ERX9 A0A0K8SKG2 A0A1B6CCS9 A0A2S2PHU9 T1PBD9 A0A091KT94 Q59FQ4 B3M317 A0A1I8NSV1 A0A1L8EEM7 V9ICN1 A0A3B0JQU1 P54385-2 A0A1W4UJV6 I5ANX6 B4QSD4 Q298M7 B4G4C1 A0A3B0KCM3 T1P987 B4PL48 B3P8B7 E2C3C7 B4N9E8 B4HG10 A0A1S4FQT2 P54385 B4M0C5 A0A2U3YY02 A0A3Q0E0N0 A0A1L8EER7 A0A0Q9WUT7 A0A1I8NSU8 A0A2S2PUP3 P00367-3 B4K740 T1HLZ6
PDB
6G2U
E-value=3.9998e-22,
Score=252
Ontologies
PATHWAY
00220
Arginine biosynthesis - Bombyx mori (domestic silkworm)
00250 Alanine, aspartate and glutamate metabolism - Bombyx mori (domestic silkworm)
00471 D-Glutamine and D-glutamate metabolism - Bombyx mori (domestic silkworm)
00910 Nitrogen metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
00250 Alanine, aspartate and glutamate metabolism - Bombyx mori (domestic silkworm)
00471 D-Glutamine and D-glutamate metabolism - Bombyx mori (domestic silkworm)
00910 Nitrogen metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
GO
GO:0016491
GO:0006520
GO:0016639
GO:0000166
GO:0047952
GO:0004352
GO:0004367
GO:0070728
GO:0005739
GO:0006537
GO:0043531
GO:0005525
GO:0006538
GO:0005759
GO:0004353
GO:0006116
GO:0042802
GO:0006536
GO:0005524
GO:0005829
GO:0005737
GO:0019551
GO:0070403
GO:0032024
GO:0006541
GO:0008652
GO:0072350
GO:0021762
GO:0009226
GO:0005515
GO:0051103
GO:0008270
GO:0042555
GO:0048384
GO:0055114
Topology
Subcellular location
Mitochondrion matrix
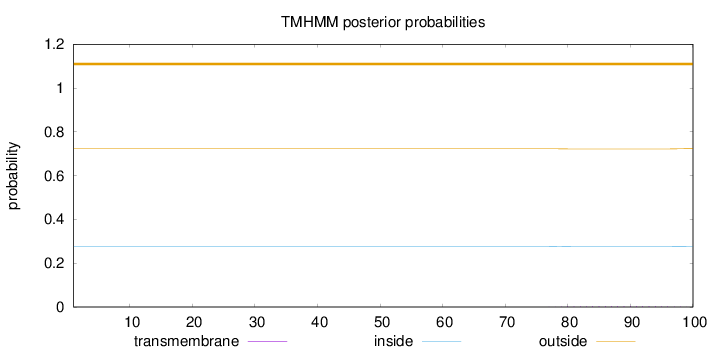
Length:
100
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.06553
Exp number, first 60 AAs:
0.00022
Total prob of N-in:
0.27703
outside
1 - 100
Population Genetic Test Statistics
Pi
12.604882
Theta
12.973646
Tajima's D
-0.413063
CLR
0.66258
CSRT
0.266586670666467
Interpretation
Uncertain
