Gene
KWMTBOMO03689
Pre Gene Modal
BGIBMGA014491
Annotation
PREDICTED:_glutamate_dehydrogenase?_mitochondrial-like_[Bombyx_mori]
Full name
Glutamate dehydrogenase
Location in the cell
Cytoplasmic Reliability : 1.869 Mitochondrial Reliability : 2.256
Sequence
CDS
ATGACCATCAGTAACTTTATTCACAACGAGCATTGGATGAAACAGATCGGCTTGAAGCCCGGCTTCGAGGGCAAAACAGCTATTATTCAAGGTTACGGCAACGTGGGCAGTTTTGCTGCCATTTACCTTCAGCAGCACGGAGTCAAAGTCATCGGGGTTTTAGAAGCTGATTGCAACCTCCACAAACCTGAAGGGTTAAACACTGTGGAGTTACTGCAGTACAAACAGAAGAACAAGGGAAGCGCTAAAGGCTACCCGAACGCCAAAGACGTGGGCCCGGATTTGTTATACGAAAACTGCGACATCCTCGTCGTCGCGGCGATGGAGAAGACTCTCTCACAACAGGTCGCAGCCAAACTGAACTGTAAGATTATCGGCGAAGGCGCGAACGGTCCAACGACTCCGGCCGCGGACGTGATACTCCGCGACAAGAAAGTCCTGGTCATCCCGGACTTGCTAGCGAACGCGGGCGGAGTCACCGTCTCGTACTTCGAATTCTTGAAGAATATCAACCATGTCAGCTTCGGGAAATTGAGTATCAAGTTCTGGCGAGACTCTAATACTGCATTACTAGATTCCGTTGAAAAGTCGTTAAAAGCGGCCAATATCGACGCGAAAATGGGACCCACGCCGCTGTTCAAGTCGCTGATGTCCGGCGCCAACGAGGAGCACATTGTCAACTCTGGGCTCGAGTACTCTATGACCAACGCTTGTAAAGTACGTCAACTGTCATTCGTTGGGTGA
Protein
MTISNFIHNEHWMKQIGLKPGFEGKTAIIQGYGNVGSFAAIYLQQHGVKVIGVLEADCNLHKPEGLNTVELLQYKQKNKGSAKGYPNAKDVGPDLLYENCDILVVAAMEKTLSQQVAAKLNCKIIGEGANGPTTPAADVILRDKKVLVIPDLLANAGGVTVSYFEFLKNINHVSFGKLSIKFWRDSNTALLDSVEKSLKAANIDAKMGPTPLFKSLMSGANEEHIVNSGLEYSMTNACKVRQLSFVG
Summary
Similarity
Belongs to the Glu/Leu/Phe/Val dehydrogenases family.
Uniprot
A0A194RE92
A0A194QHH4
A0A2A4JI69
A0A212FKL6
A0A3S2M4V8
A0A2H1WUM6
+ More
A0A0L7LFF5 A0A183UMT8 A0A0B2UXI4 F1L1P6 A0A0M3ISM5 E0VD85 A0A0J9Y6S1 A0A044T0R5 A0A0R3QQH1 T2MH20 A0A1S0UE31 A0A3P7DLL1 A0A0K0JSW1 A0A0N4TSG0 A0A183I9C8 A0A183VB84 A0A1I7W421 A0A0R3RUG8 Q0IG14 A0A1S4F5X6 D6WCS6 A0A3P6TKV7 A0A0N5CM30 A0A183E9N4 A0A2C9K6L1 A0A182TI31 A0A182VPR7 A0A182REA3 A0A182TAN9 A0A182M4A2 A0A182H069 A0A3S3SHV3 A0A182JD22 A0A182PSR1 A0A0N5AJQ7 A0A182IDH1 A0A182YES6 A0A1W4W2V6 A0A0B2VUX1 A0A182NDZ6 T1EAG6 A0A182XDR2 F1L1D2 A0A182QPR0 A0A182JVX8 A0A1B6F9V0 Q7PPE1 A0A1B6ERL0 A0A183B287 A0A0R3TNF8 A0A0Q9X1Y5 A0A1I8EYJ3 A0A0M3QYL6 A0A0X3PYS3 A0A183T5E0 A0A146NJ20 A0A1Q3FSX3 A0A0N5A8V8 A0A0N4UZV9 A0A0N4UEX9 A0A3G5ANZ7 T1JVD9 A0A1D2MF77 A0A1Q3FT11 B4K740 A0A0Q9WR91 A0A2M3ZZI4 A0A3M7PMT4 A0A2G9UD45 A0A183VC48 A0A267FVZ7 A0A0N4VLS1 A0A267EFG5 A0A0P5P4Y5 A0A147B756 A0A1I8CCC2 R7V8Z9 C1LJB0 A0A2R7W1V0 A0A1Q3FJJ7 A0A1Q3FJB7 A0A2M4CT81 B0X5C8 F5HLL7 A0A1I8ASW0 A0A0P5M5H4 A0A0N8CPB4 A0A2M3YYJ1 B4M0C5 A0A2M3YYN3 A0A0C4DHE7 A0A182SSZ4 A0A0P5WBZ1
A0A0L7LFF5 A0A183UMT8 A0A0B2UXI4 F1L1P6 A0A0M3ISM5 E0VD85 A0A0J9Y6S1 A0A044T0R5 A0A0R3QQH1 T2MH20 A0A1S0UE31 A0A3P7DLL1 A0A0K0JSW1 A0A0N4TSG0 A0A183I9C8 A0A183VB84 A0A1I7W421 A0A0R3RUG8 Q0IG14 A0A1S4F5X6 D6WCS6 A0A3P6TKV7 A0A0N5CM30 A0A183E9N4 A0A2C9K6L1 A0A182TI31 A0A182VPR7 A0A182REA3 A0A182TAN9 A0A182M4A2 A0A182H069 A0A3S3SHV3 A0A182JD22 A0A182PSR1 A0A0N5AJQ7 A0A182IDH1 A0A182YES6 A0A1W4W2V6 A0A0B2VUX1 A0A182NDZ6 T1EAG6 A0A182XDR2 F1L1D2 A0A182QPR0 A0A182JVX8 A0A1B6F9V0 Q7PPE1 A0A1B6ERL0 A0A183B287 A0A0R3TNF8 A0A0Q9X1Y5 A0A1I8EYJ3 A0A0M3QYL6 A0A0X3PYS3 A0A183T5E0 A0A146NJ20 A0A1Q3FSX3 A0A0N5A8V8 A0A0N4UZV9 A0A0N4UEX9 A0A3G5ANZ7 T1JVD9 A0A1D2MF77 A0A1Q3FT11 B4K740 A0A0Q9WR91 A0A2M3ZZI4 A0A3M7PMT4 A0A2G9UD45 A0A183VC48 A0A267FVZ7 A0A0N4VLS1 A0A267EFG5 A0A0P5P4Y5 A0A147B756 A0A1I8CCC2 R7V8Z9 C1LJB0 A0A2R7W1V0 A0A1Q3FJJ7 A0A1Q3FJB7 A0A2M4CT81 B0X5C8 F5HLL7 A0A1I8ASW0 A0A0P5M5H4 A0A0N8CPB4 A0A2M3YYJ1 B4M0C5 A0A2M3YYN3 A0A0C4DHE7 A0A182SSZ4 A0A0P5WBZ1
Pubmed
EMBL
KQ460500
KPJ14246.1
KQ459011
KPJ04385.1
NWSH01001373
PCG71489.1
+ More
AGBW02008029 OWR54281.1 RSAL01000033 RVE51459.1 ODYU01011146 SOQ56707.1 JTDY01001348 KOB74114.1 UYWY01020291 VDM41129.1 JPKZ01003059 KHN73782.1 JI169691 ADY44050.1 DS235072 EEB11341.1 LN855219 CDQ03230.1 CMVM020000121 UZAG01016196 VDO26617.1 HAAD01005326 CDG71558.1 JH712563 EJD73849.1 UYWW01001621 VDM10770.1 CDQ03229.1 UZAD01013238 VDN92777.1 UZAM01000410 VDO80971.1 UYWY01025025 VDM49325.1 CH477281 EAT44909.1 KQ971311 EEZ99054.2 UYRX01000559 VDK83843.1 UYYF01000134 VDM96497.1 UYRT01085526 VDN30244.1 AXCM01000238 JXUM01021986 KQ560617 KXJ81598.1 NCKU01000444 NCKU01000443 NCKU01000442 RWS15477.1 RWS15490.1 RWS15508.1 APCN01005979 JPKZ01000852 KHN85152.1 GAMD01001053 JAB00538.1 JI169507 ADY43936.1 AXCN02002212 GECZ01022779 JAS46990.1 AAAB01008952 EAA10575.6 GECZ01029180 JAS40589.1 UZAN01054813 VDP90594.1 UZAE01012427 VDO05106.1 CH933806 KRG02059.1 CP012526 ALC47893.1 GEEE01009453 JAP53772.1 UYSU01036726 VDL98073.1 GCES01154448 JAQ31874.1 GFDL01004380 JAV30665.1 UXUI01007466 VDD87744.1 UYYG01000010 VDN50925.1 MH990517 AYV89064.1 CAEY01000792 LJIJ01001474 ODM91626.1 GFDL01004306 JAV30739.1 EDW16353.1 CH940650 KRF83230.1 GGFK01000665 MBW33986.1 REGN01009982 RMZ99957.1 KZ347234 PIO68157.1 UYWY01025382 VDM49639.1 NIVC01000714 PAA77951.1 UXUI01011593 VDD96366.1 NIVC01002256 PAA59502.1 GDIQ01143569 JAL08157.1 GEIB01001777 JAR86594.1 AMQN01004639 KB294061 ELU15049.1 FN319060 CAX74788.1 KK854250 PTY13716.1 GFDL01007350 JAV27695.1 GFDL01007368 JAV27677.1 GGFL01004352 MBW68530.1 DS232374 EDS40813.1 AAAB01008898 EGK97150.1 GDIQ01160360 JAK91365.1 GDIP01112135 JAL91579.1 GGFM01000592 MBW21343.1 EDW67287.1 GGFM01000621 MBW21372.1 AE014297 AAN13963.3 GDIP01088245 JAM15470.1
AGBW02008029 OWR54281.1 RSAL01000033 RVE51459.1 ODYU01011146 SOQ56707.1 JTDY01001348 KOB74114.1 UYWY01020291 VDM41129.1 JPKZ01003059 KHN73782.1 JI169691 ADY44050.1 DS235072 EEB11341.1 LN855219 CDQ03230.1 CMVM020000121 UZAG01016196 VDO26617.1 HAAD01005326 CDG71558.1 JH712563 EJD73849.1 UYWW01001621 VDM10770.1 CDQ03229.1 UZAD01013238 VDN92777.1 UZAM01000410 VDO80971.1 UYWY01025025 VDM49325.1 CH477281 EAT44909.1 KQ971311 EEZ99054.2 UYRX01000559 VDK83843.1 UYYF01000134 VDM96497.1 UYRT01085526 VDN30244.1 AXCM01000238 JXUM01021986 KQ560617 KXJ81598.1 NCKU01000444 NCKU01000443 NCKU01000442 RWS15477.1 RWS15490.1 RWS15508.1 APCN01005979 JPKZ01000852 KHN85152.1 GAMD01001053 JAB00538.1 JI169507 ADY43936.1 AXCN02002212 GECZ01022779 JAS46990.1 AAAB01008952 EAA10575.6 GECZ01029180 JAS40589.1 UZAN01054813 VDP90594.1 UZAE01012427 VDO05106.1 CH933806 KRG02059.1 CP012526 ALC47893.1 GEEE01009453 JAP53772.1 UYSU01036726 VDL98073.1 GCES01154448 JAQ31874.1 GFDL01004380 JAV30665.1 UXUI01007466 VDD87744.1 UYYG01000010 VDN50925.1 MH990517 AYV89064.1 CAEY01000792 LJIJ01001474 ODM91626.1 GFDL01004306 JAV30739.1 EDW16353.1 CH940650 KRF83230.1 GGFK01000665 MBW33986.1 REGN01009982 RMZ99957.1 KZ347234 PIO68157.1 UYWY01025382 VDM49639.1 NIVC01000714 PAA77951.1 UXUI01011593 VDD96366.1 NIVC01002256 PAA59502.1 GDIQ01143569 JAL08157.1 GEIB01001777 JAR86594.1 AMQN01004639 KB294061 ELU15049.1 FN319060 CAX74788.1 KK854250 PTY13716.1 GFDL01007350 JAV27695.1 GFDL01007368 JAV27677.1 GGFL01004352 MBW68530.1 DS232374 EDS40813.1 AAAB01008898 EGK97150.1 GDIQ01160360 JAK91365.1 GDIP01112135 JAL91579.1 GGFM01000592 MBW21343.1 EDW67287.1 GGFM01000621 MBW21372.1 AE014297 AAN13963.3 GDIP01088245 JAM15470.1
Proteomes
UP000053240
UP000053268
UP000218220
UP000007151
UP000283053
UP000037510
+ More
UP000050794 UP000267007 UP000031036 UP000036681 UP000009046 UP000006672 UP000024404 UP000050602 UP000270924 UP000038020 UP000278627 UP000050793 UP000095285 UP000050640 UP000008820 UP000007266 UP000277928 UP000046394 UP000276776 UP000050760 UP000076420 UP000075902 UP000075920 UP000075900 UP000075901 UP000075883 UP000069940 UP000249989 UP000285301 UP000075880 UP000075885 UP000046393 UP000075840 UP000076408 UP000192223 UP000075884 UP000076407 UP000075886 UP000075881 UP000007062 UP000050740 UP000046398 UP000278807 UP000009192 UP000093561 UP000092553 UP000050788 UP000275846 UP000038041 UP000274131 UP000038040 UP000274756 UP000015104 UP000094527 UP000008792 UP000276133 UP000215902 UP000095286 UP000014760 UP000002320 UP000095287 UP000000803
UP000050794 UP000267007 UP000031036 UP000036681 UP000009046 UP000006672 UP000024404 UP000050602 UP000270924 UP000038020 UP000278627 UP000050793 UP000095285 UP000050640 UP000008820 UP000007266 UP000277928 UP000046394 UP000276776 UP000050760 UP000076420 UP000075902 UP000075920 UP000075900 UP000075901 UP000075883 UP000069940 UP000249989 UP000285301 UP000075880 UP000075885 UP000046393 UP000075840 UP000076408 UP000192223 UP000075884 UP000076407 UP000075886 UP000075881 UP000007062 UP000050740 UP000046398 UP000278807 UP000009192 UP000093561 UP000092553 UP000050788 UP000275846 UP000038041 UP000274131 UP000038040 UP000274756 UP000015104 UP000094527 UP000008792 UP000276133 UP000215902 UP000095286 UP000014760 UP000002320 UP000095287 UP000000803
Interpro
SUPFAM
SSF51735
SSF51735
ProteinModelPortal
A0A194RE92
A0A194QHH4
A0A2A4JI69
A0A212FKL6
A0A3S2M4V8
A0A2H1WUM6
+ More
A0A0L7LFF5 A0A183UMT8 A0A0B2UXI4 F1L1P6 A0A0M3ISM5 E0VD85 A0A0J9Y6S1 A0A044T0R5 A0A0R3QQH1 T2MH20 A0A1S0UE31 A0A3P7DLL1 A0A0K0JSW1 A0A0N4TSG0 A0A183I9C8 A0A183VB84 A0A1I7W421 A0A0R3RUG8 Q0IG14 A0A1S4F5X6 D6WCS6 A0A3P6TKV7 A0A0N5CM30 A0A183E9N4 A0A2C9K6L1 A0A182TI31 A0A182VPR7 A0A182REA3 A0A182TAN9 A0A182M4A2 A0A182H069 A0A3S3SHV3 A0A182JD22 A0A182PSR1 A0A0N5AJQ7 A0A182IDH1 A0A182YES6 A0A1W4W2V6 A0A0B2VUX1 A0A182NDZ6 T1EAG6 A0A182XDR2 F1L1D2 A0A182QPR0 A0A182JVX8 A0A1B6F9V0 Q7PPE1 A0A1B6ERL0 A0A183B287 A0A0R3TNF8 A0A0Q9X1Y5 A0A1I8EYJ3 A0A0M3QYL6 A0A0X3PYS3 A0A183T5E0 A0A146NJ20 A0A1Q3FSX3 A0A0N5A8V8 A0A0N4UZV9 A0A0N4UEX9 A0A3G5ANZ7 T1JVD9 A0A1D2MF77 A0A1Q3FT11 B4K740 A0A0Q9WR91 A0A2M3ZZI4 A0A3M7PMT4 A0A2G9UD45 A0A183VC48 A0A267FVZ7 A0A0N4VLS1 A0A267EFG5 A0A0P5P4Y5 A0A147B756 A0A1I8CCC2 R7V8Z9 C1LJB0 A0A2R7W1V0 A0A1Q3FJJ7 A0A1Q3FJB7 A0A2M4CT81 B0X5C8 F5HLL7 A0A1I8ASW0 A0A0P5M5H4 A0A0N8CPB4 A0A2M3YYJ1 B4M0C5 A0A2M3YYN3 A0A0C4DHE7 A0A182SSZ4 A0A0P5WBZ1
A0A0L7LFF5 A0A183UMT8 A0A0B2UXI4 F1L1P6 A0A0M3ISM5 E0VD85 A0A0J9Y6S1 A0A044T0R5 A0A0R3QQH1 T2MH20 A0A1S0UE31 A0A3P7DLL1 A0A0K0JSW1 A0A0N4TSG0 A0A183I9C8 A0A183VB84 A0A1I7W421 A0A0R3RUG8 Q0IG14 A0A1S4F5X6 D6WCS6 A0A3P6TKV7 A0A0N5CM30 A0A183E9N4 A0A2C9K6L1 A0A182TI31 A0A182VPR7 A0A182REA3 A0A182TAN9 A0A182M4A2 A0A182H069 A0A3S3SHV3 A0A182JD22 A0A182PSR1 A0A0N5AJQ7 A0A182IDH1 A0A182YES6 A0A1W4W2V6 A0A0B2VUX1 A0A182NDZ6 T1EAG6 A0A182XDR2 F1L1D2 A0A182QPR0 A0A182JVX8 A0A1B6F9V0 Q7PPE1 A0A1B6ERL0 A0A183B287 A0A0R3TNF8 A0A0Q9X1Y5 A0A1I8EYJ3 A0A0M3QYL6 A0A0X3PYS3 A0A183T5E0 A0A146NJ20 A0A1Q3FSX3 A0A0N5A8V8 A0A0N4UZV9 A0A0N4UEX9 A0A3G5ANZ7 T1JVD9 A0A1D2MF77 A0A1Q3FT11 B4K740 A0A0Q9WR91 A0A2M3ZZI4 A0A3M7PMT4 A0A2G9UD45 A0A183VC48 A0A267FVZ7 A0A0N4VLS1 A0A267EFG5 A0A0P5P4Y5 A0A147B756 A0A1I8CCC2 R7V8Z9 C1LJB0 A0A2R7W1V0 A0A1Q3FJJ7 A0A1Q3FJB7 A0A2M4CT81 B0X5C8 F5HLL7 A0A1I8ASW0 A0A0P5M5H4 A0A0N8CPB4 A0A2M3YYJ1 B4M0C5 A0A2M3YYN3 A0A0C4DHE7 A0A182SSZ4 A0A0P5WBZ1
PDB
1L1F
E-value=6.71711e-56,
Score=547
Ontologies
PATHWAY
00220
Arginine biosynthesis - Bombyx mori (domestic silkworm)
00250 Alanine, aspartate and glutamate metabolism - Bombyx mori (domestic silkworm)
00471 D-Glutamine and D-glutamate metabolism - Bombyx mori (domestic silkworm)
00910 Nitrogen metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
00250 Alanine, aspartate and glutamate metabolism - Bombyx mori (domestic silkworm)
00471 D-Glutamine and D-glutamate metabolism - Bombyx mori (domestic silkworm)
00910 Nitrogen metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
GO
Topology
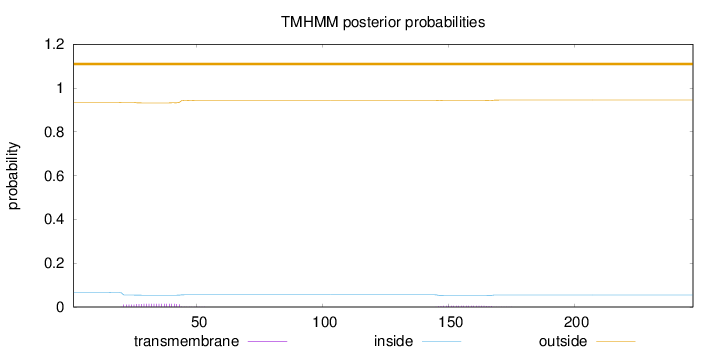
Length:
247
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.43027
Exp number, first 60 AAs:
0.31051
Total prob of N-in:
0.06566
outside
1 - 247
Population Genetic Test Statistics
Pi
4.460472
Theta
15.587816
Tajima's D
-1.397914
CLR
0.179132
CSRT
0.0709464526773661
Interpretation
Uncertain
