Gene
KWMTBOMO03688 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA014492
Annotation
putative_nad_dependent_epimerase/dehydratase_[Danaus_plexippus]
Location in the cell
Cytoplasmic Reliability : 1.694 Nuclear Reliability : 1.116
Sequence
CDS
ATGAAATTAAATGGATTTAAACGAGGTTCTCAAGAGGGCAAGAAGCAACGTCGATTTATAACTATAAAGATGAGTGACGGAAATGTTGTTCTTGTGACCGGAGGCTCCGGACTAGTTGGTCAAGCAATAAAGACAGTTATAGAGCGCGACAGACAGAAACGAAATAGTGATTATGATAGTGAGACGTGGATATTTAGCGGTTCCAAGGATGGCGACCTCCGAGATAAAACTCAAACAGAAGCTCTATTTGCTAAACACAAGCCTACCCATGTAATACATTTAGCGGCAATGGTTGGGGGACTGTTTCACAATATGGCACACAATTTGGATTTCTTTAGAGAGAATATGTCAATAAATGATAATATTCTACAAGCGTGTCACAAATATAATGTAAAGAAGGTTGTATCTTGTCTCTCGACATGCATATTTCCTGATAAGACAACATATCCTATTGATGAGACTATGGTACACAATGGTCCACCTCATAGCTCCAATTTTGGATATAGCTATGCCAAAAGAATGATTGACGTTTTAAATCGAGGATACAATGAGTCTTATGGTTGTATGTTCACCTCAGTTATTCCTTGTAATGTGTTTGGGCCATACGACAACTTCAGTTTAGAATCCAGTCATGTAATACCGGCTCTTATAAGGAGGATGGATGATGCTATGCAAAAAGGTGATCCCACGTTCACAGTGATGGGAAGCGGCAAACCACTGCGACAGTTTATTTATTCATTAGATCTAGCCGAGCTGTTCATTTGGGTCCTCAGGAACTACAACAGTATCGAACCTATTATCTTATCAGTTGACGAAAAAGACGAAGTAACGATAAGCAGAGTGGCGGAGATGATTAAGCGAGCCCACGGATACGGCGGAGATATTGTCTATGACACGACCAGAGCCGACGGCCAGTACAAGAAGACCGCGTCCAACACTAAACTCAGAACTTTGTATGAAGATTTCGAATTTACGCCATTCGATAAAGCCATAGACGAGACTGTCAAATGGTTTAAATTGAATAAAAATAGCGCTAGACTATAA
Protein
MKLNGFKRGSQEGKKQRRFITIKMSDGNVVLVTGGSGLVGQAIKTVIERDRQKRNSDYDSETWIFSGSKDGDLRDKTQTEALFAKHKPTHVIHLAAMVGGLFHNMAHNLDFFRENMSINDNILQACHKYNVKKVVSCLSTCIFPDKTTYPIDETMVHNGPPHSSNFGYSYAKRMIDVLNRGYNESYGCMFTSVIPCNVFGPYDNFSLESSHVIPALIRRMDDAMQKGDPTFTVMGSGKPLRQFIYSLDLAELFIWVLRNYNSIEPIILSVDEKDEVTISRVAEMIKRAHGYGGDIVYDTTRADGQYKKTASNTKLRTLYEDFEFTPFDKAIDETVKWFKLNKNSARL
Summary
Uniprot
H9JY74
A0A2H1WUG7
A0A437BMI2
A0A2A4JI91
A0A212F867
A0A194QG44
+ More
A0A2P8XQP5 A0A0N1I645 A0A2J7Q241 A0A067R980 A0A1B6MEY9 A0A1B6CZH8 E0W157 A0A154PK74 A0A2S2PXT9 D2A3S2 J3JXU2 A0A1B6F6Q5 A0A1S4EPN4 A0A146L414 A0A0M8ZX91 N6U1T5 K7J7J2 U4UI27 A0A0J7KPN6 A0A0L7R069 R4FQ79 A0A1W4WDM5 E2C9C1 A0A0P4VTX7 A0A026X2L2 A0A224XTX6 A0A1Y1LFZ0 A0A088AKI9 A0A336MDA6 A0A2A3EKW6 A0A182RTC6 F4W7F3 U5EW51 J9K3X5 C4WW44 A0A182L1K0 A0A336MFY6 A0A182LSJ5 A0A433QTR7 A0A151X8R0 A0A068S1Y7 A0A195DSN2 A0A151JSS2 A0A158NPU1 A0A1X2GZD1 A0A0C9QQ84 A0A195ATF3 A0A182WU56 A0A0B1TQS6 A0A182QA08 A0A182UK82 Q7Q3X9 E9IX38 A0A182Y056 A0A151I6V4 A0A433DGD1 A0A077WGJ2 A0A182HUR9 A0A023ENI9 A0A182V755 A0A182G4M7 A0A1Y1Y642 A0A182JYH6 A0A1Y1YY80 B4MJL3 A0A182PMN3 A0A182SVJ3 A0A182W757 F1LB29 A0A182GRM3 A0A3R7MB09 A0A2R7X266 A0A023F8E2 A0A162PUT5 B0W4F6 A0A1I8PTG2 A0A0C9MS07 Q16YA4 A0A084WFZ4 Q16FS9 A0A1Q3FFD6 A0A182J9T8 A0A1X2HR39 A0A2M4BTY2 A0A0C2CWK6 B4P8M0 A0A168J0R6 A0A433P8C3 A0A131YNE4 A0A224YXP5 A0A1E1XI47 S2JTA4 A0A2M4BUC0 C1BU94
A0A2P8XQP5 A0A0N1I645 A0A2J7Q241 A0A067R980 A0A1B6MEY9 A0A1B6CZH8 E0W157 A0A154PK74 A0A2S2PXT9 D2A3S2 J3JXU2 A0A1B6F6Q5 A0A1S4EPN4 A0A146L414 A0A0M8ZX91 N6U1T5 K7J7J2 U4UI27 A0A0J7KPN6 A0A0L7R069 R4FQ79 A0A1W4WDM5 E2C9C1 A0A0P4VTX7 A0A026X2L2 A0A224XTX6 A0A1Y1LFZ0 A0A088AKI9 A0A336MDA6 A0A2A3EKW6 A0A182RTC6 F4W7F3 U5EW51 J9K3X5 C4WW44 A0A182L1K0 A0A336MFY6 A0A182LSJ5 A0A433QTR7 A0A151X8R0 A0A068S1Y7 A0A195DSN2 A0A151JSS2 A0A158NPU1 A0A1X2GZD1 A0A0C9QQ84 A0A195ATF3 A0A182WU56 A0A0B1TQS6 A0A182QA08 A0A182UK82 Q7Q3X9 E9IX38 A0A182Y056 A0A151I6V4 A0A433DGD1 A0A077WGJ2 A0A182HUR9 A0A023ENI9 A0A182V755 A0A182G4M7 A0A1Y1Y642 A0A182JYH6 A0A1Y1YY80 B4MJL3 A0A182PMN3 A0A182SVJ3 A0A182W757 F1LB29 A0A182GRM3 A0A3R7MB09 A0A2R7X266 A0A023F8E2 A0A162PUT5 B0W4F6 A0A1I8PTG2 A0A0C9MS07 Q16YA4 A0A084WFZ4 Q16FS9 A0A1Q3FFD6 A0A182J9T8 A0A1X2HR39 A0A2M4BTY2 A0A0C2CWK6 B4P8M0 A0A168J0R6 A0A433P8C3 A0A131YNE4 A0A224YXP5 A0A1E1XI47 S2JTA4 A0A2M4BUC0 C1BU94
Pubmed
19121390
22118469
26354079
29403074
24845553
20566863
+ More
18362917 19820115 22516182 26823975 23537049 20075255 20798317 27129103 24508170 30249741 28004739 21719571 20966253 30485448 21347285 12364791 14747013 17210077 21282665 25244985 24945155 26483478 17994087 21685128 25474469 17510324 24438588 17550304 26830274 28797301 28503490
18362917 19820115 22516182 26823975 23537049 20075255 20798317 27129103 24508170 30249741 28004739 21719571 20966253 30485448 21347285 12364791 14747013 17210077 21282665 25244985 24945155 26483478 17994087 21685128 25474469 17510324 24438588 17550304 26830274 28797301 28503490
EMBL
BABH01042679
ODYU01011146
SOQ56708.1
RSAL01000033
RVE51458.1
NWSH01001373
+ More
PCG71486.1 AGBW02009779 OWR49935.1 KQ459011 KPJ04384.1 PYGN01001516 PSN34327.1 KQ458522 KPJ05844.1 NEVH01019373 PNF22653.1 KK852805 KDR16135.1 GEBQ01005484 JAT34493.1 GEDC01018438 JAS18860.1 DS235866 EEB19363.1 KQ434943 KZC12256.1 GGMS01000977 MBY70180.1 KQ971348 EFA04892.1 BT128063 AEE63024.1 GECZ01023851 JAS45918.1 GDHC01017109 JAQ01520.1 KQ435808 KOX72980.1 APGK01052894 KB741216 ENN72517.1 KB632366 ERL93634.1 LBMM01004537 KMQ92312.1 KQ414672 KOC64222.1 ACPB03021815 GAHY01000559 JAA76951.1 GL453806 EFN75466.1 GDKW01001141 JAI55454.1 KK107021 QOIP01000011 EZA62328.1 RLU17002.1 GFTR01004997 JAW11429.1 GEZM01056965 GEZM01056964 JAV72543.1 UFQT01000745 SSX26889.1 KZ288217 PBC32367.1 GL887844 EGI69826.1 GANO01001579 JAB58292.1 ABLF02024681 AK341842 BAH72114.1 UFQS01001173 UFQT01001173 SSX09382.1 SSX29284.1 AXCM01007134 RBNJ01001419 RUS33180.1 KQ982409 KYQ56719.1 CBTN010000033 CDH55862.1 KQ980487 KYN15925.1 KQ982014 KYN30639.1 ADTU01022754 MCGN01000014 ORY89550.1 GBYB01005849 JAG75616.1 KQ976745 KYM75471.1 KN549353 KHJ98137.1 AXCN02000770 AAAB01008964 EAA12379.3 GL766616 EFZ14908.1 KQ978456 KYM93889.1 RBNI01001849 RUP49911.1 LK023321 CDS06741.1 APCN01002475 GAPW01002621 JAC10977.1 JXUM01144060 KQ569713 KXJ68536.1 MCFE01000233 ORX93500.1 MCFE01000051 ORY02988.1 CH963846 EDW72302.1 JI176029 ADY47333.1 JXUM01082743 JXUM01082744 JXUM01082745 KQ563334 KXJ74033.1 QCYY01001463 ROT77871.1 KK856492 PTY25894.1 GBBI01001142 JAC17570.1 KV440979 OAD74216.1 DS231837 EDS33539.1 DF836401 GAN06112.1 CH477519 EAT39623.1 ATLV01023414 KE525343 KFB49138.1 CH478383 EAT33092.1 GFDL01008792 JAV26253.1 MCGE01000056 ORZ01917.1 GGFJ01007399 MBW56540.1 KN739401 KIH54262.1 CM000158 EDW92237.1 AMYB01000007 OAD00604.1 RBNK01008187 RUS13786.1 GEDV01008469 JAP80088.1 GFPF01007947 MAA19093.1 GFAC01000517 JAT98671.1 KE124088 EPB83015.1 GGFJ01007400 MBW56541.1 BT078173 BT120592 HACA01025463 ACO12597.1 ADD24232.1 CDW42824.1
PCG71486.1 AGBW02009779 OWR49935.1 KQ459011 KPJ04384.1 PYGN01001516 PSN34327.1 KQ458522 KPJ05844.1 NEVH01019373 PNF22653.1 KK852805 KDR16135.1 GEBQ01005484 JAT34493.1 GEDC01018438 JAS18860.1 DS235866 EEB19363.1 KQ434943 KZC12256.1 GGMS01000977 MBY70180.1 KQ971348 EFA04892.1 BT128063 AEE63024.1 GECZ01023851 JAS45918.1 GDHC01017109 JAQ01520.1 KQ435808 KOX72980.1 APGK01052894 KB741216 ENN72517.1 KB632366 ERL93634.1 LBMM01004537 KMQ92312.1 KQ414672 KOC64222.1 ACPB03021815 GAHY01000559 JAA76951.1 GL453806 EFN75466.1 GDKW01001141 JAI55454.1 KK107021 QOIP01000011 EZA62328.1 RLU17002.1 GFTR01004997 JAW11429.1 GEZM01056965 GEZM01056964 JAV72543.1 UFQT01000745 SSX26889.1 KZ288217 PBC32367.1 GL887844 EGI69826.1 GANO01001579 JAB58292.1 ABLF02024681 AK341842 BAH72114.1 UFQS01001173 UFQT01001173 SSX09382.1 SSX29284.1 AXCM01007134 RBNJ01001419 RUS33180.1 KQ982409 KYQ56719.1 CBTN010000033 CDH55862.1 KQ980487 KYN15925.1 KQ982014 KYN30639.1 ADTU01022754 MCGN01000014 ORY89550.1 GBYB01005849 JAG75616.1 KQ976745 KYM75471.1 KN549353 KHJ98137.1 AXCN02000770 AAAB01008964 EAA12379.3 GL766616 EFZ14908.1 KQ978456 KYM93889.1 RBNI01001849 RUP49911.1 LK023321 CDS06741.1 APCN01002475 GAPW01002621 JAC10977.1 JXUM01144060 KQ569713 KXJ68536.1 MCFE01000233 ORX93500.1 MCFE01000051 ORY02988.1 CH963846 EDW72302.1 JI176029 ADY47333.1 JXUM01082743 JXUM01082744 JXUM01082745 KQ563334 KXJ74033.1 QCYY01001463 ROT77871.1 KK856492 PTY25894.1 GBBI01001142 JAC17570.1 KV440979 OAD74216.1 DS231837 EDS33539.1 DF836401 GAN06112.1 CH477519 EAT39623.1 ATLV01023414 KE525343 KFB49138.1 CH478383 EAT33092.1 GFDL01008792 JAV26253.1 MCGE01000056 ORZ01917.1 GGFJ01007399 MBW56540.1 KN739401 KIH54262.1 CM000158 EDW92237.1 AMYB01000007 OAD00604.1 RBNK01008187 RUS13786.1 GEDV01008469 JAP80088.1 GFPF01007947 MAA19093.1 GFAC01000517 JAT98671.1 KE124088 EPB83015.1 GGFJ01007400 MBW56541.1 BT078173 BT120592 HACA01025463 ACO12597.1 ADD24232.1 CDW42824.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000007151
UP000053268
UP000245037
+ More
UP000235965 UP000027135 UP000009046 UP000076502 UP000007266 UP000079169 UP000053105 UP000019118 UP000002358 UP000030742 UP000036403 UP000053825 UP000015103 UP000192223 UP000008237 UP000053097 UP000279307 UP000005203 UP000242457 UP000075900 UP000007755 UP000007819 UP000075882 UP000075883 UP000075809 UP000027586 UP000078492 UP000078541 UP000005205 UP000242180 UP000078540 UP000076407 UP000075886 UP000075902 UP000007062 UP000076408 UP000078542 UP000075840 UP000075903 UP000069940 UP000249989 UP000193498 UP000075881 UP000007798 UP000075885 UP000075901 UP000075920 UP000283509 UP000077315 UP000002320 UP000095300 UP000053815 UP000008820 UP000030765 UP000075880 UP000193560 UP000002282 UP000077051 UP000014254
UP000235965 UP000027135 UP000009046 UP000076502 UP000007266 UP000079169 UP000053105 UP000019118 UP000002358 UP000030742 UP000036403 UP000053825 UP000015103 UP000192223 UP000008237 UP000053097 UP000279307 UP000005203 UP000242457 UP000075900 UP000007755 UP000007819 UP000075882 UP000075883 UP000075809 UP000027586 UP000078492 UP000078541 UP000005205 UP000242180 UP000078540 UP000076407 UP000075886 UP000075902 UP000007062 UP000076408 UP000078542 UP000075840 UP000075903 UP000069940 UP000249989 UP000193498 UP000075881 UP000007798 UP000075885 UP000075901 UP000075920 UP000283509 UP000077315 UP000002320 UP000095300 UP000053815 UP000008820 UP000030765 UP000075880 UP000193560 UP000002282 UP000077051 UP000014254
PRIDE
Pfam
PF01370 Epimerase
Interpro
SUPFAM
SSF51735
SSF51735
ProteinModelPortal
H9JY74
A0A2H1WUG7
A0A437BMI2
A0A2A4JI91
A0A212F867
A0A194QG44
+ More
A0A2P8XQP5 A0A0N1I645 A0A2J7Q241 A0A067R980 A0A1B6MEY9 A0A1B6CZH8 E0W157 A0A154PK74 A0A2S2PXT9 D2A3S2 J3JXU2 A0A1B6F6Q5 A0A1S4EPN4 A0A146L414 A0A0M8ZX91 N6U1T5 K7J7J2 U4UI27 A0A0J7KPN6 A0A0L7R069 R4FQ79 A0A1W4WDM5 E2C9C1 A0A0P4VTX7 A0A026X2L2 A0A224XTX6 A0A1Y1LFZ0 A0A088AKI9 A0A336MDA6 A0A2A3EKW6 A0A182RTC6 F4W7F3 U5EW51 J9K3X5 C4WW44 A0A182L1K0 A0A336MFY6 A0A182LSJ5 A0A433QTR7 A0A151X8R0 A0A068S1Y7 A0A195DSN2 A0A151JSS2 A0A158NPU1 A0A1X2GZD1 A0A0C9QQ84 A0A195ATF3 A0A182WU56 A0A0B1TQS6 A0A182QA08 A0A182UK82 Q7Q3X9 E9IX38 A0A182Y056 A0A151I6V4 A0A433DGD1 A0A077WGJ2 A0A182HUR9 A0A023ENI9 A0A182V755 A0A182G4M7 A0A1Y1Y642 A0A182JYH6 A0A1Y1YY80 B4MJL3 A0A182PMN3 A0A182SVJ3 A0A182W757 F1LB29 A0A182GRM3 A0A3R7MB09 A0A2R7X266 A0A023F8E2 A0A162PUT5 B0W4F6 A0A1I8PTG2 A0A0C9MS07 Q16YA4 A0A084WFZ4 Q16FS9 A0A1Q3FFD6 A0A182J9T8 A0A1X2HR39 A0A2M4BTY2 A0A0C2CWK6 B4P8M0 A0A168J0R6 A0A433P8C3 A0A131YNE4 A0A224YXP5 A0A1E1XI47 S2JTA4 A0A2M4BUC0 C1BU94
A0A2P8XQP5 A0A0N1I645 A0A2J7Q241 A0A067R980 A0A1B6MEY9 A0A1B6CZH8 E0W157 A0A154PK74 A0A2S2PXT9 D2A3S2 J3JXU2 A0A1B6F6Q5 A0A1S4EPN4 A0A146L414 A0A0M8ZX91 N6U1T5 K7J7J2 U4UI27 A0A0J7KPN6 A0A0L7R069 R4FQ79 A0A1W4WDM5 E2C9C1 A0A0P4VTX7 A0A026X2L2 A0A224XTX6 A0A1Y1LFZ0 A0A088AKI9 A0A336MDA6 A0A2A3EKW6 A0A182RTC6 F4W7F3 U5EW51 J9K3X5 C4WW44 A0A182L1K0 A0A336MFY6 A0A182LSJ5 A0A433QTR7 A0A151X8R0 A0A068S1Y7 A0A195DSN2 A0A151JSS2 A0A158NPU1 A0A1X2GZD1 A0A0C9QQ84 A0A195ATF3 A0A182WU56 A0A0B1TQS6 A0A182QA08 A0A182UK82 Q7Q3X9 E9IX38 A0A182Y056 A0A151I6V4 A0A433DGD1 A0A077WGJ2 A0A182HUR9 A0A023ENI9 A0A182V755 A0A182G4M7 A0A1Y1Y642 A0A182JYH6 A0A1Y1YY80 B4MJL3 A0A182PMN3 A0A182SVJ3 A0A182W757 F1LB29 A0A182GRM3 A0A3R7MB09 A0A2R7X266 A0A023F8E2 A0A162PUT5 B0W4F6 A0A1I8PTG2 A0A0C9MS07 Q16YA4 A0A084WFZ4 Q16FS9 A0A1Q3FFD6 A0A182J9T8 A0A1X2HR39 A0A2M4BTY2 A0A0C2CWK6 B4P8M0 A0A168J0R6 A0A433P8C3 A0A131YNE4 A0A224YXP5 A0A1E1XI47 S2JTA4 A0A2M4BUC0 C1BU94
PDB
4E5Y
E-value=1.81799e-110,
Score=1020
Ontologies
PATHWAY
GO
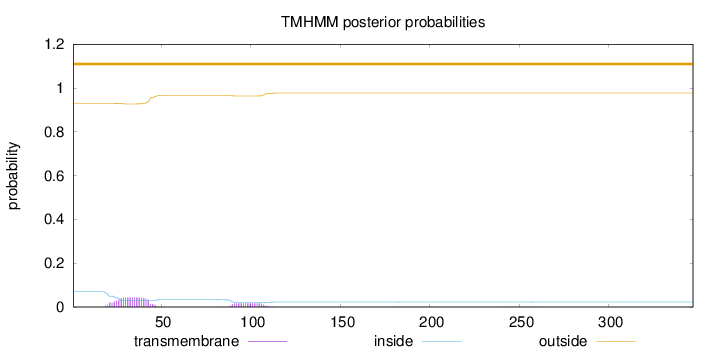
Topology
Length:
347
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.24983
Exp number, first 60 AAs:
0.90499
Total prob of N-in:
0.06920
outside
1 - 347
Population Genetic Test Statistics
Pi
3.451985
Theta
13.535649
Tajima's D
-0.85177
CLR
0.533223
CSRT
0.165841707914604
Interpretation
Uncertain
