Gene
KWMTBOMO03684
Annotation
PREDICTED:_protein_transport_protein_SFT2_[Plutella_xylostella]
Full name
Vesicle transport protein
Location in the cell
PlasmaMembrane Reliability : 4.831
Sequence
CDS
ATGGCAAATCTAAAATCAGATTTAGATCAATATTTGCTTCAAAACGAAAGTCGTCGGAGTTATAAATTTAGTTTACCAAGTTTTTCAACGCCTGCCTTCCTGTCTCGTAACACTGAGGAGACTCCGACGAGCACAACGAGCAGTTGGTTCGAAGAAGTACAGAAAGAGTATTTCACTTTGAGCAGAACACAACGATTCTTAGGATTTGGAATATGTCTATTTCTAGGAATCCTATGTTTCATACTATCATTTATCTACATACCAGTTCTTCTATTACAAGCTAGGAAATTTGCTCTACTGTTCACATTGGGAAGTTTATTCTTTATACTTAGTTTCAGCTTTCTCTACGGACCATGGGCACATTTGAAGTCAATGTTCTCAAAGGAAAGAGCTCTTACAACATCAATTTACAGTATAACTTTGATAGCGACATTATACTGTGCCTTACATCTTCAAAGTACACCGTGGACCATAGTATGTGCTGTCTTACAAGTCATGGCATTGTTCTGGATGATGATGGGGTCTATACCAGGAGGATCATCGGGCATGAGGTTCTTTGGAAGCATGTTCAAATCATCAGTTTCAAACACATTGCCGATTTAA
Protein
MANLKSDLDQYLLQNESRRSYKFSLPSFSTPAFLSRNTEETPTSTTSSWFEEVQKEYFTLSRTQRFLGFGICLFLGILCFILSFIYIPVLLLQARKFALLFTLGSLFFILSFSFLYGPWAHLKSMFSKERALTTSIYSITLIATLYCALHLQSTPWTIVCAVLQVMALFWMMMGSIPGGSSGMRFFGSMFKSSVSNTLPI
Summary
Description
May be involved in fusion of retrograde transport vesicles derived from an endocytic compartment with the Golgi complex.
Similarity
Belongs to the SFT2 family.
Uniprot
A0A194RAE7
A0A1E1WDR2
S4PYG3
A0A194QLQ8
A0A2H1WCP2
A0A2A4JXG4
+ More
A0A3S2PHA8 A0A212F4B4 D6WYV8 A0A1W4XQX0 A0A1Y1KBC2 A0A0T6AVM9 J3JYX2 A0A0L7RIX7 A0A2J7RQR9 A0A088AVN6 A0A2A3EK70 A0A0M9A527 A0A154PE65 A0A026WYA4 A0A067RPM5 A0A2P8ZNS9 A0A1B6KXL4 K7IRJ7 A0A3B0J9L0 A0A0V0GCT4 A0A0P4VSW6 R4G8M5 E2AGK8 A0A069DQ21 A0A023F999 A0A158P1S9 A0A0A9XJV9 B4P2A4 A0A224XKH8 A0A336MWH0 Q177W9 A0A2M4AKH4 A0A151J8M6 T1IC97 W5JMC6 U5EU38 A0A195ERF3 A0A1I8NMN1 B5DI31 B4G8F4 B0X4G7 A0A0L0CD12 A0A084VPD5 A0A1Q3FGL4 B4ICT0 B4Q5N4 Q6II06 A0A023EIA8 A8E703 B3MUC7 A0A151XAN7 B4MTV1 A8E718 B3N785 A0A182J1A8 A0A2M4BZU0 A0A1I8NI80 T1PKL8 A0A2M3Z9S9 A0A1W4V2R3 A0A034WJW2 A0A0K8VN95 A0A182NED7 A0A182Q9N7 Q7QG63 A0A182S4Q9 A0A182L5P2 A0A182MK60 A0A182V3E5 A0A182VV04 J9JZK0 A0A182IE28 A0A1B0CFN6 A0A1L8DUX7 A0A1B6CFX2 A0A182K822
A0A3S2PHA8 A0A212F4B4 D6WYV8 A0A1W4XQX0 A0A1Y1KBC2 A0A0T6AVM9 J3JYX2 A0A0L7RIX7 A0A2J7RQR9 A0A088AVN6 A0A2A3EK70 A0A0M9A527 A0A154PE65 A0A026WYA4 A0A067RPM5 A0A2P8ZNS9 A0A1B6KXL4 K7IRJ7 A0A3B0J9L0 A0A0V0GCT4 A0A0P4VSW6 R4G8M5 E2AGK8 A0A069DQ21 A0A023F999 A0A158P1S9 A0A0A9XJV9 B4P2A4 A0A224XKH8 A0A336MWH0 Q177W9 A0A2M4AKH4 A0A151J8M6 T1IC97 W5JMC6 U5EU38 A0A195ERF3 A0A1I8NMN1 B5DI31 B4G8F4 B0X4G7 A0A0L0CD12 A0A084VPD5 A0A1Q3FGL4 B4ICT0 B4Q5N4 Q6II06 A0A023EIA8 A8E703 B3MUC7 A0A151XAN7 B4MTV1 A8E718 B3N785 A0A182J1A8 A0A2M4BZU0 A0A1I8NI80 T1PKL8 A0A2M3Z9S9 A0A1W4V2R3 A0A034WJW2 A0A0K8VN95 A0A182NED7 A0A182Q9N7 Q7QG63 A0A182S4Q9 A0A182L5P2 A0A182MK60 A0A182V3E5 A0A182VV04 J9JZK0 A0A182IE28 A0A1B0CFN6 A0A1L8DUX7 A0A1B6CFX2 A0A182K822
Pubmed
26354079
23622113
22118469
18362917
19820115
28004739
+ More
22516182 23537049 24508170 30249741 24845553 29403074 20075255 27129103 20798317 26334808 25474469 21347285 25401762 26823975 17994087 17550304 17510324 20920257 23761445 15632085 26108605 24438588 22936249 10731132 12537568 12537572 12537573 12537574 14709175 16110336 17569856 17569867 26109357 26109356 24945155 26483478 25315136 25348373 12364791 14747013 17210077 20966253
22516182 23537049 24508170 30249741 24845553 29403074 20075255 27129103 20798317 26334808 25474469 21347285 25401762 26823975 17994087 17550304 17510324 20920257 23761445 15632085 26108605 24438588 22936249 10731132 12537568 12537572 12537573 12537574 14709175 16110336 17569856 17569867 26109357 26109356 24945155 26483478 25315136 25348373 12364791 14747013 17210077 20966253
EMBL
KQ460500
KPJ14250.1
GDQN01005930
JAT85124.1
GAIX01003338
JAA89222.1
+ More
KQ459011 KPJ04381.1 ODYU01007761 SOQ50807.1 NWSH01000430 PCG76466.1 RSAL01000033 RVE51455.1 AGBW02010402 OWR48575.1 KQ971363 EFA09012.1 GEZM01087262 JAV58812.1 LJIG01022692 KRT79219.1 APGK01057128 BT128453 KB741277 KB632243 AEE63410.1 ENN71295.1 ERL90552.1 KQ414582 KOC70815.1 NEVH01000618 PNF43182.1 KZ288220 PBC32203.1 KQ435745 KOX76616.1 KQ434874 KZC09714.1 KK107069 QOIP01000002 EZA60726.1 RLU25979.1 KK852544 KDR21649.1 PYGN01000008 PSN58167.1 GEBQ01023805 JAT16172.1 OUUW01000004 SPP78954.1 GECL01000236 JAP05888.1 GDKW01000329 JAI56266.1 GAHY01000712 JAA76798.1 GL439322 EFN67428.1 GBGD01002959 JAC85930.1 GBBI01000636 JAC18076.1 ADTU01006781 GBHO01023668 GBRD01001100 GDHC01019158 JAG19936.1 JAG64721.1 JAP99470.1 CM000157 EDW87100.1 GFTR01003441 JAW12985.1 UFQT01001383 SSX30308.1 CH477370 EAT42456.1 GGFK01007962 MBW41283.1 KQ979492 KYN21213.1 ACPB03027240 ADMH02001016 ETN64448.1 GANO01004054 JAB55817.1 KQ982021 KYN30482.1 CH379060 EDY69969.1 CH479180 EDW28634.1 DS232340 EDS40320.1 JRES01000673 KNC29354.1 ATLV01014998 KE524999 KFB39829.1 GFDL01008356 JAV26689.1 CH480829 EDW45356.1 CM000361 CM002910 EDX03184.1 KMY87255.1 AE014134 BK003260 AAZ66445.1 DAA03459.1 JXUM01013126 GAPW01004465 KQ560369 JAC09133.1 KXJ82757.1 BT030945 ABV82327.1 CH902624 EDV33456.1 KQ982335 KYQ57442.1 CH963852 EDW75540.1 BT030960 ABV82342.1 CH954177 EDV57192.1 GGFJ01009444 MBW58585.1 KA648478 AFP63107.1 GGFM01004548 MBW25299.1 GAKP01003091 JAC55861.1 GDHF01011960 JAI40354.1 AXCN02000860 AAAB01008839 EAA05881.3 AXCM01007640 ABLF02023678 APCN01004556 AJWK01010201 GFDF01003847 JAV10237.1 GEDC01025063 GEDC01000536 JAS12235.1 JAS36762.1
KQ459011 KPJ04381.1 ODYU01007761 SOQ50807.1 NWSH01000430 PCG76466.1 RSAL01000033 RVE51455.1 AGBW02010402 OWR48575.1 KQ971363 EFA09012.1 GEZM01087262 JAV58812.1 LJIG01022692 KRT79219.1 APGK01057128 BT128453 KB741277 KB632243 AEE63410.1 ENN71295.1 ERL90552.1 KQ414582 KOC70815.1 NEVH01000618 PNF43182.1 KZ288220 PBC32203.1 KQ435745 KOX76616.1 KQ434874 KZC09714.1 KK107069 QOIP01000002 EZA60726.1 RLU25979.1 KK852544 KDR21649.1 PYGN01000008 PSN58167.1 GEBQ01023805 JAT16172.1 OUUW01000004 SPP78954.1 GECL01000236 JAP05888.1 GDKW01000329 JAI56266.1 GAHY01000712 JAA76798.1 GL439322 EFN67428.1 GBGD01002959 JAC85930.1 GBBI01000636 JAC18076.1 ADTU01006781 GBHO01023668 GBRD01001100 GDHC01019158 JAG19936.1 JAG64721.1 JAP99470.1 CM000157 EDW87100.1 GFTR01003441 JAW12985.1 UFQT01001383 SSX30308.1 CH477370 EAT42456.1 GGFK01007962 MBW41283.1 KQ979492 KYN21213.1 ACPB03027240 ADMH02001016 ETN64448.1 GANO01004054 JAB55817.1 KQ982021 KYN30482.1 CH379060 EDY69969.1 CH479180 EDW28634.1 DS232340 EDS40320.1 JRES01000673 KNC29354.1 ATLV01014998 KE524999 KFB39829.1 GFDL01008356 JAV26689.1 CH480829 EDW45356.1 CM000361 CM002910 EDX03184.1 KMY87255.1 AE014134 BK003260 AAZ66445.1 DAA03459.1 JXUM01013126 GAPW01004465 KQ560369 JAC09133.1 KXJ82757.1 BT030945 ABV82327.1 CH902624 EDV33456.1 KQ982335 KYQ57442.1 CH963852 EDW75540.1 BT030960 ABV82342.1 CH954177 EDV57192.1 GGFJ01009444 MBW58585.1 KA648478 AFP63107.1 GGFM01004548 MBW25299.1 GAKP01003091 JAC55861.1 GDHF01011960 JAI40354.1 AXCN02000860 AAAB01008839 EAA05881.3 AXCM01007640 ABLF02023678 APCN01004556 AJWK01010201 GFDF01003847 JAV10237.1 GEDC01025063 GEDC01000536 JAS12235.1 JAS36762.1
Proteomes
UP000053240
UP000053268
UP000218220
UP000283053
UP000007151
UP000007266
+ More
UP000192223 UP000019118 UP000030742 UP000053825 UP000235965 UP000005203 UP000242457 UP000053105 UP000076502 UP000053097 UP000279307 UP000027135 UP000245037 UP000002358 UP000268350 UP000000311 UP000005205 UP000002282 UP000008820 UP000078492 UP000015103 UP000000673 UP000078541 UP000095300 UP000001819 UP000008744 UP000002320 UP000037069 UP000030765 UP000001292 UP000000304 UP000000803 UP000069940 UP000249989 UP000007801 UP000075809 UP000007798 UP000008711 UP000075880 UP000095301 UP000192221 UP000075884 UP000075886 UP000007062 UP000075900 UP000075882 UP000075883 UP000075903 UP000075920 UP000007819 UP000075840 UP000092461 UP000075881
UP000192223 UP000019118 UP000030742 UP000053825 UP000235965 UP000005203 UP000242457 UP000053105 UP000076502 UP000053097 UP000279307 UP000027135 UP000245037 UP000002358 UP000268350 UP000000311 UP000005205 UP000002282 UP000008820 UP000078492 UP000015103 UP000000673 UP000078541 UP000095300 UP000001819 UP000008744 UP000002320 UP000037069 UP000030765 UP000001292 UP000000304 UP000000803 UP000069940 UP000249989 UP000007801 UP000075809 UP000007798 UP000008711 UP000075880 UP000095301 UP000192221 UP000075884 UP000075886 UP000007062 UP000075900 UP000075882 UP000075883 UP000075903 UP000075920 UP000007819 UP000075840 UP000092461 UP000075881
PRIDE
Interpro
CDD
ProteinModelPortal
A0A194RAE7
A0A1E1WDR2
S4PYG3
A0A194QLQ8
A0A2H1WCP2
A0A2A4JXG4
+ More
A0A3S2PHA8 A0A212F4B4 D6WYV8 A0A1W4XQX0 A0A1Y1KBC2 A0A0T6AVM9 J3JYX2 A0A0L7RIX7 A0A2J7RQR9 A0A088AVN6 A0A2A3EK70 A0A0M9A527 A0A154PE65 A0A026WYA4 A0A067RPM5 A0A2P8ZNS9 A0A1B6KXL4 K7IRJ7 A0A3B0J9L0 A0A0V0GCT4 A0A0P4VSW6 R4G8M5 E2AGK8 A0A069DQ21 A0A023F999 A0A158P1S9 A0A0A9XJV9 B4P2A4 A0A224XKH8 A0A336MWH0 Q177W9 A0A2M4AKH4 A0A151J8M6 T1IC97 W5JMC6 U5EU38 A0A195ERF3 A0A1I8NMN1 B5DI31 B4G8F4 B0X4G7 A0A0L0CD12 A0A084VPD5 A0A1Q3FGL4 B4ICT0 B4Q5N4 Q6II06 A0A023EIA8 A8E703 B3MUC7 A0A151XAN7 B4MTV1 A8E718 B3N785 A0A182J1A8 A0A2M4BZU0 A0A1I8NI80 T1PKL8 A0A2M3Z9S9 A0A1W4V2R3 A0A034WJW2 A0A0K8VN95 A0A182NED7 A0A182Q9N7 Q7QG63 A0A182S4Q9 A0A182L5P2 A0A182MK60 A0A182V3E5 A0A182VV04 J9JZK0 A0A182IE28 A0A1B0CFN6 A0A1L8DUX7 A0A1B6CFX2 A0A182K822
A0A3S2PHA8 A0A212F4B4 D6WYV8 A0A1W4XQX0 A0A1Y1KBC2 A0A0T6AVM9 J3JYX2 A0A0L7RIX7 A0A2J7RQR9 A0A088AVN6 A0A2A3EK70 A0A0M9A527 A0A154PE65 A0A026WYA4 A0A067RPM5 A0A2P8ZNS9 A0A1B6KXL4 K7IRJ7 A0A3B0J9L0 A0A0V0GCT4 A0A0P4VSW6 R4G8M5 E2AGK8 A0A069DQ21 A0A023F999 A0A158P1S9 A0A0A9XJV9 B4P2A4 A0A224XKH8 A0A336MWH0 Q177W9 A0A2M4AKH4 A0A151J8M6 T1IC97 W5JMC6 U5EU38 A0A195ERF3 A0A1I8NMN1 B5DI31 B4G8F4 B0X4G7 A0A0L0CD12 A0A084VPD5 A0A1Q3FGL4 B4ICT0 B4Q5N4 Q6II06 A0A023EIA8 A8E703 B3MUC7 A0A151XAN7 B4MTV1 A8E718 B3N785 A0A182J1A8 A0A2M4BZU0 A0A1I8NI80 T1PKL8 A0A2M3Z9S9 A0A1W4V2R3 A0A034WJW2 A0A0K8VN95 A0A182NED7 A0A182Q9N7 Q7QG63 A0A182S4Q9 A0A182L5P2 A0A182MK60 A0A182V3E5 A0A182VV04 J9JZK0 A0A182IE28 A0A1B0CFN6 A0A1L8DUX7 A0A1B6CFX2 A0A182K822
Ontologies
GO
PANTHER
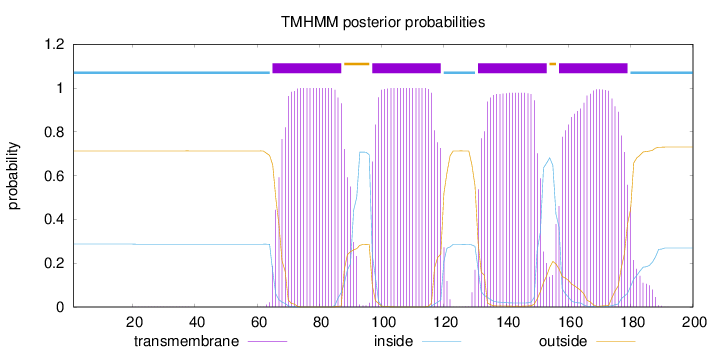
Topology
Subcellular location
Membrane
Length:
200
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
86.98918
Exp number, first 60 AAs:
0.00729
Total prob of N-in:
0.28738
inside
1 - 64
TMhelix
65 - 87
outside
88 - 96
TMhelix
97 - 119
inside
120 - 130
TMhelix
131 - 153
outside
154 - 156
TMhelix
157 - 179
inside
180 - 200
Population Genetic Test Statistics
Pi
23.75796
Theta
22.560826
Tajima's D
0.157342
CLR
1.261185
CSRT
0.419079046047698
Interpretation
Uncertain
