Gene
KWMTBOMO03674
Pre Gene Modal
BGIBMGA002214
Annotation
PREDICTED:_WD_repeat-containing_protein_44_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.959
Sequence
CDS
ATGCGGATCACGTCGGAGTACATCGGCCGGACCAACGACGACGAGACCACCAGGGAGACAGATGAAGAAAGCGAAAGCGTTATTCCGGGCGGAGCCGACGACGAAAGCAAAGAGACGAAACGCGAATCAACAGACAGACAGACAACCGGAATCAAGAGGAAAACCGAAAAGTTAAAACGTTTCTTCGGGAGCACGATGAAGAAAACCGTGGACGCGGCCAAGTCGTTGGCGCAGGAGGTGTCGCACGCGCGGCACAAGGAGGACGTGGCGGACATAGCGGACGACGTCCGCGGCGAGCACAACATCAAGCTGAAGGCCTCCAACTCGCACAAGGGGCCCTACGACTTCGACGGCTGCGTGCGTCACGTACAGGAGATGGGGTCGGGGCACTGCGGCGCGGTGTGGTGCTGCAAGTTCAGCGTGTGCGGGCGCCTGCTGGCCACGGCCGGCCAGGACCGCCTGCTGCGCGTGTGGGCCACCAGGGACGCGCACCACATGTTCCAGGACATGCGGACGAAATACAACGCGGAACAGAAGTCGTCCCCCACCCCCTCGCAAGAGTCCCTGGCGTCCCTGGCCCCCCTAGCGCCGCCCCCCGCCCCCGAAGACGCCCCCCTTGGCCCCTCCGCCCCTTTCTGTCCCAAACCGTTCTGCGTCTACTCCGGTCACACCTCCGACCTGCTGGACGTATCCTGGTCGAAGAACTACTTCATCCTGTCCAGCTCCATGGACAAGACGGTCAGGCTGTGGCACATCTCCCGGGGGGAGTGCCTGTGCTGCTTCCAGCACATTGACTTCGTCACGGCCATTGTGTTCCATCCCAGAGACGACAGGTACTTCCTGTCGGGCAGTCTGGACGGGAAGCTGAGGCTGTGGGATATACCTGATAAAAAGGTGGCGGTCTGGAACGAGGTGGACGGCAAGACGAAGCTCATCACGGCGGCCAACTTCTGCCAGAACGGCAAGTTCGCCGTCGTGGGCACGTACGACGGCCGCTGCATCTTCTACACCACCGACCAGCTCAAGTACCACACGCAGATCGACGTGCGCTCGACCAGGGGCAAGAACTCCACCGGCCGCAAGATCAGCGGCATCGAGCCCATGCCCAACGACGACAAGATCCTCGTCACGTCCAACGACAGCAGGATCCGGCTGTACGACCTGCGCGACCTCAACCTCTCGTGCAAGTACAAGGGCTACGTGAACGTGTCGAGCCAGATCAAGGCGTCGTTCTCGCACGACGGCAAGTACATTGTGAGCGGCTCGGAGAACCAGTGCATCTACATCTGGAAGACGCACCACGACTACTCCAAGTTCACGTCGGTGCGGCGCGACCGGAACGACTTCTGGGAGGGGATCAAGGCTCACAACGCGGTGGTCACGTGCGCCGTGTTCGCGCCCAACCCCGACCACATGATCCGCATGATCAACGAGCGGGAGCACGAGCTCAGGCTGGCCGGCGACGCGCCCGACCAGGAGGACGACCGCCTGAAGGGGCGCGAGACGGGGCTGGTGTCGTCCTCGCAGCCGGGCCACGTGCTGGTGAGCGCGGACTTCAACGGCTGCATCAAAGTGTTCGTGCACAAGGCCAAGCCCAAGCACAGCTCCCTCCCAGCCTCCGCGCTCGCAAATGGAAAGTCGAGCAGTCGAGCCAGTTGA
Protein
MRITSEYIGRTNDDETTRETDEESESVIPGGADDESKETKRESTDRQTTGIKRKTEKLKRFFGSTMKKTVDAAKSLAQEVSHARHKEDVADIADDVRGEHNIKLKASNSHKGPYDFDGCVRHVQEMGSGHCGAVWCCKFSVCGRLLATAGQDRLLRVWATRDAHHMFQDMRTKYNAEQKSSPTPSQESLASLAPLAPPPAPEDAPLGPSAPFCPKPFCVYSGHTSDLLDVSWSKNYFILSSSMDKTVRLWHISRGECLCCFQHIDFVTAIVFHPRDDRYFLSGSLDGKLRLWDIPDKKVAVWNEVDGKTKLITAANFCQNGKFAVVGTYDGRCIFYTTDQLKYHTQIDVRSTRGKNSTGRKISGIEPMPNDDKILVTSNDSRIRLYDLRDLNLSCKYKGYVNVSSQIKASFSHDGKYIVSGSENQCIYIWKTHHDYSKFTSVRRDRNDFWEGIKAHNAVVTCAVFAPNPDHMIRMINEREHELRLAGDAPDQEDDRLKGRETGLVSSSQPGHVLVSADFNGCIKVFVHKAKPKHSSLPASALANGKSSSRAS
Summary
Uniprot
H9IY81
A0A2A4JD75
A0A2H1WG24
A0A194QG34
A0A212EI28
A0A0N1PK43
+ More
A0A2A3EC15 K7J4S6 A0A088AES2 A0A154PKU5 A0A232EVK1 E2BP70 A0A026WTE1 F4WRC3 A0A310SST0 A0A195BAJ5 A0A0J7KNW1 A0A151J2Y0 A0A151I6H4 A0A0M9A972 A0A0C9PYM9 A0A195FJG0 A0A151WRG5 A0A2J7QG27 A0A2P8XE00 E9JCH5 A0A158NUU7 E2AZA5 A0A0L0CIS6 I5AMW3 T1PCF0 A0A1I8PEC9 A0A1I8PEL5 A0A1I8MJP4 B4GNJ8 A0A3B0K5S9 Q0KHZ4 B4K6J9 A0A0C4DHB8 D0QWP9 B4NKB1 B3LYT9 Q8IMK1 A0A0M4F7V4 A0A0Q9WUY5 B4M5Y5 A0A0T6B5S2 B4R0U9 Q7K501 B4PPI0 B4HZH1 A0A1Q3FA64 U5EUK3 A0A0R1E4L6 A0A1Q3FA55 B0WZY4 A0A1B0FEP0 B3P5B1 Q16HY0 A0A1S4G088 A0A1Y1L648 A0A182GA32 A0A182GY22 A0A1A9Y252 A0A1A9V5Z4 A0A1B0BGV9 A0A1A9ZEA5 A0A0L7QZE0 B4JU32 A0A1W4URS1 A0A1W4UDB8 A0A3Q0IHS2 A0A3B0JF50 A0A0L7KV62 D6WEM4 V5GX89 A0A0K8U5X2 A0A0K8VJR8 W8BKB4 A0A0K8U272 A0A0K8UDZ8 V5H0M9 A0A1B0CE02 A0A336M7T3 A0A336KKK6 A0A336M9V6 A0A1A9W2T0 A0A1L8DNK9 E0VKN1 A0A182K6E5 U4UBN7 A0A1B6M199 A0A0A1XGY5 A0A182PVA3 W8BWY3 A0A182WYA4 A0A1B6FDJ7 A0A182HIG7 Q7PRV0 A0A182N1D8
A0A2A3EC15 K7J4S6 A0A088AES2 A0A154PKU5 A0A232EVK1 E2BP70 A0A026WTE1 F4WRC3 A0A310SST0 A0A195BAJ5 A0A0J7KNW1 A0A151J2Y0 A0A151I6H4 A0A0M9A972 A0A0C9PYM9 A0A195FJG0 A0A151WRG5 A0A2J7QG27 A0A2P8XE00 E9JCH5 A0A158NUU7 E2AZA5 A0A0L0CIS6 I5AMW3 T1PCF0 A0A1I8PEC9 A0A1I8PEL5 A0A1I8MJP4 B4GNJ8 A0A3B0K5S9 Q0KHZ4 B4K6J9 A0A0C4DHB8 D0QWP9 B4NKB1 B3LYT9 Q8IMK1 A0A0M4F7V4 A0A0Q9WUY5 B4M5Y5 A0A0T6B5S2 B4R0U9 Q7K501 B4PPI0 B4HZH1 A0A1Q3FA64 U5EUK3 A0A0R1E4L6 A0A1Q3FA55 B0WZY4 A0A1B0FEP0 B3P5B1 Q16HY0 A0A1S4G088 A0A1Y1L648 A0A182GA32 A0A182GY22 A0A1A9Y252 A0A1A9V5Z4 A0A1B0BGV9 A0A1A9ZEA5 A0A0L7QZE0 B4JU32 A0A1W4URS1 A0A1W4UDB8 A0A3Q0IHS2 A0A3B0JF50 A0A0L7KV62 D6WEM4 V5GX89 A0A0K8U5X2 A0A0K8VJR8 W8BKB4 A0A0K8U272 A0A0K8UDZ8 V5H0M9 A0A1B0CE02 A0A336M7T3 A0A336KKK6 A0A336M9V6 A0A1A9W2T0 A0A1L8DNK9 E0VKN1 A0A182K6E5 U4UBN7 A0A1B6M199 A0A0A1XGY5 A0A182PVA3 W8BWY3 A0A182WYA4 A0A1B6FDJ7 A0A182HIG7 Q7PRV0 A0A182N1D8
Pubmed
19121390
26354079
22118469
20075255
28648823
20798317
+ More
24508170 30249741 21719571 29403074 21282665 21347285 26108605 15632085 17994087 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 19859648 18057021 26109357 26109356 17550304 17510324 28004739 26483478 26227816 18362917 19820115 24495485 20566863 23537049 25830018 12364791 14747013 17210077
24508170 30249741 21719571 29403074 21282665 21347285 26108605 15632085 17994087 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 19859648 18057021 26109357 26109356 17550304 17510324 28004739 26483478 26227816 18362917 19820115 24495485 20566863 23537049 25830018 12364791 14747013 17210077
EMBL
BABH01038367
NWSH01001884
PCG69799.1
ODYU01008429
SOQ52029.1
KQ459011
+ More
KPJ04374.1 AGBW02014704 OWR41163.1 KQ459677 KPJ20480.1 KZ288301 PBC28832.1 AAZX01007335 KQ434948 KZC12475.1 NNAY01001991 OXU22367.1 GL449553 EFN82490.1 KK107109 QOIP01000008 EZA59330.1 RLU19875.1 GL888284 EGI63340.1 KQ760545 OAD59970.1 KQ976532 KYM81571.1 LBMM01004995 KMQ91929.1 KQ980314 KYN16681.1 KQ978473 KYM93717.1 KQ435710 KOX79780.1 GBYB01006633 JAG76400.1 KQ981512 KYN40800.1 KQ982806 KYQ50440.1 NEVH01014836 PNF27521.1 PYGN01002583 PSN30230.1 GL771831 EFZ09476.1 ADTU01026674 ADTU01026675 GL444149 EFN61248.1 JRES01000338 KNC32151.1 CM000070 EIM52298.1 KA646436 AFP61065.1 CH479186 EDW39324.1 OUUW01000005 SPP80966.1 AE014297 ABI31217.1 CH933806 EDW16299.2 AAF56961.3 FJ821034 ACN94764.1 CH964272 EDW84041.2 CH902617 EDV44055.2 KPU80598.1 KPU80599.1 BT030455 AAN14200.3 ABP87897.1 CP012526 ALC48007.1 KRF99950.1 CH940652 EDW59061.2 LJIG01009640 KRT82676.1 CM000364 EDX14920.1 AY051546 AAK92970.1 CM000160 EDW98230.2 CH480819 EDW53428.1 GFDL01010584 JAV24461.1 GANO01003843 JAB56028.1 KRK04168.1 GFDL01010581 JAV24464.1 DS232221 EDS37809.1 CCAG010005922 CH954182 EDV53161.1 CH478130 EAT33868.1 GEZM01065953 GEZM01065952 GEZM01065950 GEZM01065947 JAV68281.1 JXUM01050177 JXUM01050178 JXUM01050179 KQ561637 KXJ77922.1 JXUM01020975 JXUM01020976 KQ560587 KXJ81777.1 JXJN01014017 KQ414681 KOC63994.1 CH916374 EDV91002.1 SPP80967.1 JTDY01005518 KOB66919.1 KQ971326 EEZ99884.1 GALX01002069 JAB66397.1 GDHF01030358 JAI21956.1 GDHF01013197 JAI39117.1 GAMC01012844 JAB93711.1 GDHF01031537 JAI20777.1 GDHF01027412 JAI24902.1 GALX01002068 JAB66398.1 AJWK01008456 UFQT01000647 SSX26090.1 UFQS01000567 UFQT01000567 SSX05019.1 SSX25381.1 SSX26091.1 GFDF01006139 JAV07945.1 DS235250 EEB13937.1 KB632237 ERL90472.1 GEBQ01010274 GEBQ01001747 JAT29703.1 JAT38230.1 GBXI01007781 GBXI01004539 GBXI01000376 JAD06511.1 JAD09753.1 JAD13916.1 GAMC01012846 JAB93709.1 GECZ01021473 JAS48296.1 APCN01003966 AAAB01008846 EAA06543.6
KPJ04374.1 AGBW02014704 OWR41163.1 KQ459677 KPJ20480.1 KZ288301 PBC28832.1 AAZX01007335 KQ434948 KZC12475.1 NNAY01001991 OXU22367.1 GL449553 EFN82490.1 KK107109 QOIP01000008 EZA59330.1 RLU19875.1 GL888284 EGI63340.1 KQ760545 OAD59970.1 KQ976532 KYM81571.1 LBMM01004995 KMQ91929.1 KQ980314 KYN16681.1 KQ978473 KYM93717.1 KQ435710 KOX79780.1 GBYB01006633 JAG76400.1 KQ981512 KYN40800.1 KQ982806 KYQ50440.1 NEVH01014836 PNF27521.1 PYGN01002583 PSN30230.1 GL771831 EFZ09476.1 ADTU01026674 ADTU01026675 GL444149 EFN61248.1 JRES01000338 KNC32151.1 CM000070 EIM52298.1 KA646436 AFP61065.1 CH479186 EDW39324.1 OUUW01000005 SPP80966.1 AE014297 ABI31217.1 CH933806 EDW16299.2 AAF56961.3 FJ821034 ACN94764.1 CH964272 EDW84041.2 CH902617 EDV44055.2 KPU80598.1 KPU80599.1 BT030455 AAN14200.3 ABP87897.1 CP012526 ALC48007.1 KRF99950.1 CH940652 EDW59061.2 LJIG01009640 KRT82676.1 CM000364 EDX14920.1 AY051546 AAK92970.1 CM000160 EDW98230.2 CH480819 EDW53428.1 GFDL01010584 JAV24461.1 GANO01003843 JAB56028.1 KRK04168.1 GFDL01010581 JAV24464.1 DS232221 EDS37809.1 CCAG010005922 CH954182 EDV53161.1 CH478130 EAT33868.1 GEZM01065953 GEZM01065952 GEZM01065950 GEZM01065947 JAV68281.1 JXUM01050177 JXUM01050178 JXUM01050179 KQ561637 KXJ77922.1 JXUM01020975 JXUM01020976 KQ560587 KXJ81777.1 JXJN01014017 KQ414681 KOC63994.1 CH916374 EDV91002.1 SPP80967.1 JTDY01005518 KOB66919.1 KQ971326 EEZ99884.1 GALX01002069 JAB66397.1 GDHF01030358 JAI21956.1 GDHF01013197 JAI39117.1 GAMC01012844 JAB93711.1 GDHF01031537 JAI20777.1 GDHF01027412 JAI24902.1 GALX01002068 JAB66398.1 AJWK01008456 UFQT01000647 SSX26090.1 UFQS01000567 UFQT01000567 SSX05019.1 SSX25381.1 SSX26091.1 GFDF01006139 JAV07945.1 DS235250 EEB13937.1 KB632237 ERL90472.1 GEBQ01010274 GEBQ01001747 JAT29703.1 JAT38230.1 GBXI01007781 GBXI01004539 GBXI01000376 JAD06511.1 JAD09753.1 JAD13916.1 GAMC01012846 JAB93709.1 GECZ01021473 JAS48296.1 APCN01003966 AAAB01008846 EAA06543.6
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000053240
UP000242457
+ More
UP000002358 UP000005203 UP000076502 UP000215335 UP000008237 UP000053097 UP000279307 UP000007755 UP000078540 UP000036403 UP000078492 UP000078542 UP000053105 UP000078541 UP000075809 UP000235965 UP000245037 UP000005205 UP000000311 UP000037069 UP000001819 UP000095301 UP000095300 UP000008744 UP000268350 UP000000803 UP000009192 UP000007798 UP000007801 UP000092553 UP000008792 UP000000304 UP000002282 UP000001292 UP000002320 UP000092444 UP000008711 UP000008820 UP000069940 UP000249989 UP000092443 UP000078200 UP000092460 UP000092445 UP000053825 UP000001070 UP000192221 UP000079169 UP000037510 UP000007266 UP000092461 UP000091820 UP000009046 UP000075881 UP000030742 UP000075885 UP000076407 UP000075840 UP000007062 UP000075884
UP000002358 UP000005203 UP000076502 UP000215335 UP000008237 UP000053097 UP000279307 UP000007755 UP000078540 UP000036403 UP000078492 UP000078542 UP000053105 UP000078541 UP000075809 UP000235965 UP000245037 UP000005205 UP000000311 UP000037069 UP000001819 UP000095301 UP000095300 UP000008744 UP000268350 UP000000803 UP000009192 UP000007798 UP000007801 UP000092553 UP000008792 UP000000304 UP000002282 UP000001292 UP000002320 UP000092444 UP000008711 UP000008820 UP000069940 UP000249989 UP000092443 UP000078200 UP000092460 UP000092445 UP000053825 UP000001070 UP000192221 UP000079169 UP000037510 UP000007266 UP000092461 UP000091820 UP000009046 UP000075881 UP000030742 UP000075885 UP000076407 UP000075840 UP000007062 UP000075884
Pfam
PF00400 WD40
Interpro
SUPFAM
SSF50978
SSF50978
Gene 3D
ProteinModelPortal
H9IY81
A0A2A4JD75
A0A2H1WG24
A0A194QG34
A0A212EI28
A0A0N1PK43
+ More
A0A2A3EC15 K7J4S6 A0A088AES2 A0A154PKU5 A0A232EVK1 E2BP70 A0A026WTE1 F4WRC3 A0A310SST0 A0A195BAJ5 A0A0J7KNW1 A0A151J2Y0 A0A151I6H4 A0A0M9A972 A0A0C9PYM9 A0A195FJG0 A0A151WRG5 A0A2J7QG27 A0A2P8XE00 E9JCH5 A0A158NUU7 E2AZA5 A0A0L0CIS6 I5AMW3 T1PCF0 A0A1I8PEC9 A0A1I8PEL5 A0A1I8MJP4 B4GNJ8 A0A3B0K5S9 Q0KHZ4 B4K6J9 A0A0C4DHB8 D0QWP9 B4NKB1 B3LYT9 Q8IMK1 A0A0M4F7V4 A0A0Q9WUY5 B4M5Y5 A0A0T6B5S2 B4R0U9 Q7K501 B4PPI0 B4HZH1 A0A1Q3FA64 U5EUK3 A0A0R1E4L6 A0A1Q3FA55 B0WZY4 A0A1B0FEP0 B3P5B1 Q16HY0 A0A1S4G088 A0A1Y1L648 A0A182GA32 A0A182GY22 A0A1A9Y252 A0A1A9V5Z4 A0A1B0BGV9 A0A1A9ZEA5 A0A0L7QZE0 B4JU32 A0A1W4URS1 A0A1W4UDB8 A0A3Q0IHS2 A0A3B0JF50 A0A0L7KV62 D6WEM4 V5GX89 A0A0K8U5X2 A0A0K8VJR8 W8BKB4 A0A0K8U272 A0A0K8UDZ8 V5H0M9 A0A1B0CE02 A0A336M7T3 A0A336KKK6 A0A336M9V6 A0A1A9W2T0 A0A1L8DNK9 E0VKN1 A0A182K6E5 U4UBN7 A0A1B6M199 A0A0A1XGY5 A0A182PVA3 W8BWY3 A0A182WYA4 A0A1B6FDJ7 A0A182HIG7 Q7PRV0 A0A182N1D8
A0A2A3EC15 K7J4S6 A0A088AES2 A0A154PKU5 A0A232EVK1 E2BP70 A0A026WTE1 F4WRC3 A0A310SST0 A0A195BAJ5 A0A0J7KNW1 A0A151J2Y0 A0A151I6H4 A0A0M9A972 A0A0C9PYM9 A0A195FJG0 A0A151WRG5 A0A2J7QG27 A0A2P8XE00 E9JCH5 A0A158NUU7 E2AZA5 A0A0L0CIS6 I5AMW3 T1PCF0 A0A1I8PEC9 A0A1I8PEL5 A0A1I8MJP4 B4GNJ8 A0A3B0K5S9 Q0KHZ4 B4K6J9 A0A0C4DHB8 D0QWP9 B4NKB1 B3LYT9 Q8IMK1 A0A0M4F7V4 A0A0Q9WUY5 B4M5Y5 A0A0T6B5S2 B4R0U9 Q7K501 B4PPI0 B4HZH1 A0A1Q3FA64 U5EUK3 A0A0R1E4L6 A0A1Q3FA55 B0WZY4 A0A1B0FEP0 B3P5B1 Q16HY0 A0A1S4G088 A0A1Y1L648 A0A182GA32 A0A182GY22 A0A1A9Y252 A0A1A9V5Z4 A0A1B0BGV9 A0A1A9ZEA5 A0A0L7QZE0 B4JU32 A0A1W4URS1 A0A1W4UDB8 A0A3Q0IHS2 A0A3B0JF50 A0A0L7KV62 D6WEM4 V5GX89 A0A0K8U5X2 A0A0K8VJR8 W8BKB4 A0A0K8U272 A0A0K8UDZ8 V5H0M9 A0A1B0CE02 A0A336M7T3 A0A336KKK6 A0A336M9V6 A0A1A9W2T0 A0A1L8DNK9 E0VKN1 A0A182K6E5 U4UBN7 A0A1B6M199 A0A0A1XGY5 A0A182PVA3 W8BWY3 A0A182WYA4 A0A1B6FDJ7 A0A182HIG7 Q7PRV0 A0A182N1D8
PDB
3N0D
E-value=3.40245e-19,
Score=235
Ontologies
GO
PANTHER
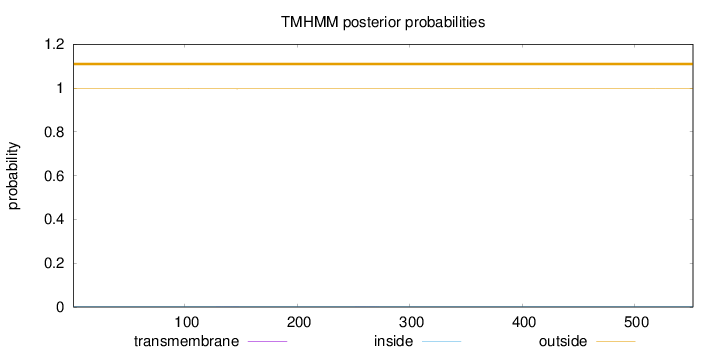
Topology
Length:
552
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01105
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00358
outside
1 - 552
Population Genetic Test Statistics
Pi
28.869188
Theta
27.595493
Tajima's D
0.234347
CLR
0.84272
CSRT
0.444827758612069
Interpretation
Uncertain
