Gene
KWMTBOMO03672
Annotation
PREDICTED:_uncharacterized_protein_LOC103314804_[Tribolium_castaneum]
Location in the cell
Nuclear Reliability : 2.029 PlasmaMembrane Reliability : 1.849
Sequence
CDS
ATGCCGCCTAAATCTAATAAATCCGAGAACGATTTGAAGCTTTTGTACGCTAAGAGGGATATGATTTATTCACGCGTGCAAACAATTTGTGATAATGCACTTAAGATATCCGACTTAGCAATACGAGACTCATTTCTTTGTTCTATTGAAAACATAGACGATTTGCGCGCGGATTTCGAAAGTATCGTTCATAATATCAACGCATTGCAATTAGAACTAGATCCTGATTACGCCGTCAGTTACTCTGTTATGTCGTCGTTTGATGATCTATATTGCCGTATAAAACGTGTCGCTAAATCTTTAGGAAAAATACCTAGCGCCGGGTATTCGCAAACGGAACCTAAGTTCACCCGCGATAGACCTAGACTACCGCCTATCCACATACCCGAATTTAATGGGGACATCCGTAATTGGGGTTTATTTATAGATAGTTTTGAAAATACAGTAAATCAAAATGATCACTTAACCGATCACGAAAAATTACACTACTTAGTAGAAAAACTTACCGGCAAAGCCCGTGCGGCTTGCGCGGGAATTACGTTTAGTGCCGAAAACTATTCTTTGATTTTGAACACACTCAAAGCCAAATATGAAGATAAACGGTTGATAGCTACCGCGTACCTGGATCAGTTGTTCGAATTAAAACCTATTTTATCGTGTACCGAACAGAATTTGGAGAAATTCTTAGACATATTTGCATCATCTGTTCGGGCTATTAGAAATCTCAACGTCAATGACCTCAGCGACTTTTTATTTTTGTACATTGGCTTAAAAAAGTTAGATTTCGAAACTGTTCGCTTATTTGAAACGAATCAGGGTACTAAGGGTATTGAATTGCCGAAATTTAATGAATTGGAATCATTTTTACGTAATCACATAAGAATACTTCAGCGTACTACGCGACCTCACGAGGTCCCGAAAAGACAACCAGAAAGGCATTTAGAAACGTCTTCAAGACCGCTTAGAACGACTCATAGCTATGTAAATACCGCGGCGTTGCCTGCGTCCGTGCCCGCCAAGTCGACTTGTTTTCTGTGCAAATCTTATCATACACAGTTGTATCAGTGTGCTGAGTTTTTGAAATTGTCGCCTAAAAATCGTTATGATTTTGTTCGCACAAATAACTTGTGTTTAAACTGCCTTAGTGATAAACATCGCGTTGGTGTGTGTAATAGTAGCCATAGTTGTCGCAAATGTTCGTTGAAGCATCATACCTTGCTTCATTTTGATAAAAATAATAATATAGTAGCCAATAGAACTACAAGCGCAGGTAATCAACCAATGTATGGTAACTCGTCGCTAGCAGTATCAAGCTCGGAGCACTCGCATGGTCGAACTCACGCTGGCGGCACCGCACGTTGTACGACACATCTCGCGCGTCCGACGTATTCACCCGCGCTTCCCGCCGCTGCTGTCGCTGCCGCTCGCACAGACAGCTCCGCAGCTATCAACTCGGACGGCAACACAAATGACGTCACATTATGCTCGCTGTCTTCCGCTCGCTTACCTACAACTGTTTTGCTTGCGACCGCTCGAGTTGTAATGGTTGATACTTATGGCAAACCGCATCTTGTTCGCGCCCTTTTAGATAGCGCCTCACAGAGTAATTTTATTACTCGTGAATGTTGTCAACGCCTTGGATTAAAATATAGTAACCAGCCTACGTGCCCCGTAGTCAAGGGCATAGGGGGGTCTACTAAGGACATCCAAGGGTCCGTAAAAATTAAATTTGAATCACGTTTTAGCTCGAATGTTTCATTCGAAATAAACTCGCTGGTTGTAGATAAAGTAACGGAGCAACTTCCTACTGTTTCTGTAGATACCTCGTTGTTAGCTAACTTTGATAGTTTGCCGTTAGCCGATGATACTTACGCCGTTCCAGGAGCCGTCGATATTTTGATTGGCGCTTCGATTTTCCCGCACCTCCTGCTGCCACGTAAGGTTCAAGGTCATTCGGCGAACCCCGCACCGCACGCTTTGGAAACTGTTCTCGGTTTTGTTATAGTTGGATCCGCACCCGCTCTTATAAATAGTTACTCGACCGTATCATACTGCTGCGCCGTCGAGCCGCCACTGGACTCCACCGTCCGCCGCTTTTGGGAAATTGAAGAAGTAAATGTTCCGCTGGTTATGAGTCCTGATGATAAGACCGCTGAGGATATTTATTGTAGCACGACCACCCGCGATGAAAGTGGTCGTTATATTGTTGCACTCCCGTTTAAGGGTGATACCTTTTCGCTTGGTGATTCGCGCCTAATGGCCGAAAAACGCTTTTTTTGTTTAGAACGTAAGATGATGGCATCTCCGAAGTTGAAAAAGGCCTACGACGATGTCGTGAATGAATATGTAGATAAGGGTATCATCTCACCTGCTCCCGCTGTACTTGATGAGACGCAAAATGGTCCCTCTTATGTCATCCCGCATCATGGTGTCATTAGGGAGGATAAACTCACCACAAAATTGCGTGTTGTTCTGGATGGCAGTGCTAAGACTAGTACTTCTGTTTCTCTAAACGACATCTTGTATTGTGGTAAAAATCTTCAAGGTAACTTGTTTGATATTATCGTCAATTTTCGTTTATTTATTGTAGCGCTATCTGCAGACATTAGACAAATGTTTCTTTGTATTGGCATTCGGGAATCGGATCGTAGATTCCAAAGATTTTTATATAGGTTTTCCCCACACGAACCATTAAAGGTTTACCAGTTTAACCGCGTTTGTTTCGGACTTAAGTCAAGTCCGTTTCACGCGCTTCGCACTGTGAAGCAGTTAGTTCTTGATGAGGGTGATTCCTACCCCAAGGCAAAAGCTACCATCAATACTGGTCTGTATATGGATGACTTTGCTTATAGTGTAGATAGCGAGCCTGAAGCCATCGAAACAGCTACTCAAGTGATAAGCCTAATGAAGACTGGGCAATTTGACCTTGTTAAGTGGACGAGCAACTCTCAACGTGTTTTGGACTGGATTCCGCCATCGCATCGTCTATCGACGGTTAAGGATTTTGATGTCAATGATACACATAAGATTCTCGGCTTGAGCTGGTCTCCCACATCCGATACATTCAGTCTCAAAATTAGCTCGACCCCGCCAACTTGTACCAAACGCACCATATTGTCTTGTGTCGCTCGCATATGGGACATTATGGGTTTTGTAGCACCTGTAGTGTTATACGCAAAGCTTCTTATTAAAGAGCTTTGGCTTTCTCAGTGTGACTGGGATGACATCGCACCGGACAACGTGGCCAGATTATGGGCTCGCTTCAGGGAAGAGCTTCCTATGCTCGAACATATAGAAATCCCACGTCATGTAGGTATCTCTCGAGATAGTGTAGTCACAATAGTGGGGTTCGCAGATGCTAGTGAGAAGGCTTATGGAGGAGTGGTGTATTTTCACGTTTGCAATGGCGACAATATTACAGTTAGCCTTATTAGTGCGAAATCTAAGGTCTCCCTCGAAAGGTCGTCTCTCTCGCGCGCCTTGAGCTTTGCGCTTTACTACTATTGA
Protein
MPPKSNKSENDLKLLYAKRDMIYSRVQTICDNALKISDLAIRDSFLCSIENIDDLRADFESIVHNINALQLELDPDYAVSYSVMSSFDDLYCRIKRVAKSLGKIPSAGYSQTEPKFTRDRPRLPPIHIPEFNGDIRNWGLFIDSFENTVNQNDHLTDHEKLHYLVEKLTGKARAACAGITFSAENYSLILNTLKAKYEDKRLIATAYLDQLFELKPILSCTEQNLEKFLDIFASSVRAIRNLNVNDLSDFLFLYIGLKKLDFETVRLFETNQGTKGIELPKFNELESFLRNHIRILQRTTRPHEVPKRQPERHLETSSRPLRTTHSYVNTAALPASVPAKSTCFLCKSYHTQLYQCAEFLKLSPKNRYDFVRTNNLCLNCLSDKHRVGVCNSSHSCRKCSLKHHTLLHFDKNNNIVANRTTSAGNQPMYGNSSLAVSSSEHSHGRTHAGGTARCTTHLARPTYSPALPAAAVAAARTDSSAAINSDGNTNDVTLCSLSSARLPTTVLLATARVVMVDTYGKPHLVRALLDSASQSNFITRECCQRLGLKYSNQPTCPVVKGIGGSTKDIQGSVKIKFESRFSSNVSFEINSLVVDKVTEQLPTVSVDTSLLANFDSLPLADDTYAVPGAVDILIGASIFPHLLLPRKVQGHSANPAPHALETVLGFVIVGSAPALINSYSTVSYCCAVEPPLDSTVRRFWEIEEVNVPLVMSPDDKTAEDIYCSTTTRDESGRYIVALPFKGDTFSLGDSRLMAEKRFFCLERKMMASPKLKKAYDDVVNEYVDKGIISPAPAVLDETQNGPSYVIPHHGVIREDKLTTKLRVVLDGSAKTSTSVSLNDILYCGKNLQGNLFDIIVNFRLFIVALSADIRQMFLCIGIRESDRRFQRFLYRFSPHEPLKVYQFNRVCFGLKSSPFHALRTVKQLVLDEGDSYPKAKATINTGLYMDDFAYSVDSEPEAIETATQVISLMKTGQFDLVKWTSNSQRVLDWIPPSHRLSTVKDFDVNDTHKILGLSWSPTSDTFSLKISSTPPTCTKRTILSCVARIWDIMGFVAPVVLYAKLLIKELWLSQCDWDDIAPDNVARLWARFREELPMLEHIEIPRHVGISRDSVVTIVGFADASEKAYGGVVYFHVCNGDNITVSLISAKSKVSLERSSLSRALSFALYYY
Summary
Uniprot
A0A3Q0JJJ7
A0A1S3DJJ0
A0A1S3DJ34
A0A1S3DJC0
A0A1Y1LIC8
A0A3Q0JFQ3
+ More
A0A3Q0JCL2 A0A1Y1LM33 A0A3Q0J7N1 A0A3Q0JBZ9 A0A3Q0J7K9 A0A3Q0J730 A0A1S3D744 A0A0J7KDN6 A0A1B6KIX6 X1XU04 J9JUK4 J9M7R2 A0A1B6MT91 X1WZF3 J9JL40 A0A0J7K7U0 X1WYQ7 A0A2S2NZ22 A0A0J7N890 A0A146KRI6 A0A1Y1LIG5 A0A1B6LLT3 A0A0A9WW84 A0A2S2P1X6 A0A1Y1KMX0 X1XU26 A0A2S2R2J0 A0A1W7R6A8 A0A1W7R6S6 A0A0A9ZHU0 A0A2S2QTX5 J9LPA9 A0A1U8N859 A0A087T9T6 A0A0J7N465 A0A2S2NHU0 X1X096 J9JLG6 J9JVZ7 A0A2H1X238 A0A1W7R6I7 A0A2S2PNZ4 A0A0N0PAG9 A0A226E780 A0A1W7R6N3 A0A226EPI5 A0A0A9WWW3 V9GZW9 J9LHB7 A0A2M4CW28 W8C1U1 A0A2M4CSC7 A0A226D417 A0A0A9X3X8
A0A3Q0JCL2 A0A1Y1LM33 A0A3Q0J7N1 A0A3Q0JBZ9 A0A3Q0J7K9 A0A3Q0J730 A0A1S3D744 A0A0J7KDN6 A0A1B6KIX6 X1XU04 J9JUK4 J9M7R2 A0A1B6MT91 X1WZF3 J9JL40 A0A0J7K7U0 X1WYQ7 A0A2S2NZ22 A0A0J7N890 A0A146KRI6 A0A1Y1LIG5 A0A1B6LLT3 A0A0A9WW84 A0A2S2P1X6 A0A1Y1KMX0 X1XU26 A0A2S2R2J0 A0A1W7R6A8 A0A1W7R6S6 A0A0A9ZHU0 A0A2S2QTX5 J9LPA9 A0A1U8N859 A0A087T9T6 A0A0J7N465 A0A2S2NHU0 X1X096 J9JLG6 J9JVZ7 A0A2H1X238 A0A1W7R6I7 A0A2S2PNZ4 A0A0N0PAG9 A0A226E780 A0A1W7R6N3 A0A226EPI5 A0A0A9WWW3 V9GZW9 J9LHB7 A0A2M4CW28 W8C1U1 A0A2M4CSC7 A0A226D417 A0A0A9X3X8
EMBL
GEZM01054789
JAV73399.1
GEZM01054766
GEZM01054765
JAV73410.1
LBMM01009202
+ More
KMQ88311.1 GEBQ01028569 JAT11408.1 ABLF02028417 ABLF02042407 ABLF02039606 ABLF02005251 ABLF02055296 GEBQ01000825 JAT39152.1 ABLF02041614 ABLF02018732 ABLF02018737 LBMM01012238 KMQ86334.1 ABLF02041670 GGMR01009860 MBY22479.1 LBMM01008464 KMQ88855.1 GDHC01019528 JAP99100.1 JAV73413.1 GEBQ01015458 JAT24519.1 GBHO01031928 GBHO01031926 JAG11676.1 JAG11678.1 GGMR01010723 MBY23342.1 GEZM01085145 GEZM01085143 JAV60187.1 ABLF02001506 ABLF02008029 ABLF02035899 ABLF02037719 ABLF02046327 ABLF02064627 GGMS01014951 MBY84154.1 GEHC01000946 JAV46699.1 GEHC01000870 JAV46775.1 GBHO01000061 JAG43543.1 GGMS01011369 MBY80572.1 ABLF02035780 KK114194 KFM61875.1 LBMM01010417 KMQ87475.1 GGMR01003717 MBY16336.1 ABLF02008032 ABLF02035293 ABLF02060940 ABLF02030698 ABLF02030705 ABLF02030709 ABLF02015138 ODYU01012875 SOQ59395.1 GEHC01000869 JAV46776.1 GGMR01018506 MBY31125.1 KQ458473 KPJ05885.1 LNIX01000005 OXA53452.1 GEHC01000865 JAV46780.1 LNIX01000002 OXA59522.1 GBHO01034255 GBHO01034254 JAG09349.1 JAG09350.1 U23420 AAB03640.1 ABLF02022040 ABLF02056888 GGFL01005358 MBW69536.1 GAMC01006864 JAB99691.1 GGFL01004049 MBW68227.1 LNIX01000036 OXA39969.1 GBHO01030122 GBHO01030121 GBHO01030120 GBHO01030116 JAG13482.1 JAG13483.1 JAG13484.1 JAG13488.1
KMQ88311.1 GEBQ01028569 JAT11408.1 ABLF02028417 ABLF02042407 ABLF02039606 ABLF02005251 ABLF02055296 GEBQ01000825 JAT39152.1 ABLF02041614 ABLF02018732 ABLF02018737 LBMM01012238 KMQ86334.1 ABLF02041670 GGMR01009860 MBY22479.1 LBMM01008464 KMQ88855.1 GDHC01019528 JAP99100.1 JAV73413.1 GEBQ01015458 JAT24519.1 GBHO01031928 GBHO01031926 JAG11676.1 JAG11678.1 GGMR01010723 MBY23342.1 GEZM01085145 GEZM01085143 JAV60187.1 ABLF02001506 ABLF02008029 ABLF02035899 ABLF02037719 ABLF02046327 ABLF02064627 GGMS01014951 MBY84154.1 GEHC01000946 JAV46699.1 GEHC01000870 JAV46775.1 GBHO01000061 JAG43543.1 GGMS01011369 MBY80572.1 ABLF02035780 KK114194 KFM61875.1 LBMM01010417 KMQ87475.1 GGMR01003717 MBY16336.1 ABLF02008032 ABLF02035293 ABLF02060940 ABLF02030698 ABLF02030705 ABLF02030709 ABLF02015138 ODYU01012875 SOQ59395.1 GEHC01000869 JAV46776.1 GGMR01018506 MBY31125.1 KQ458473 KPJ05885.1 LNIX01000005 OXA53452.1 GEHC01000865 JAV46780.1 LNIX01000002 OXA59522.1 GBHO01034255 GBHO01034254 JAG09349.1 JAG09350.1 U23420 AAB03640.1 ABLF02022040 ABLF02056888 GGFL01005358 MBW69536.1 GAMC01006864 JAB99691.1 GGFL01004049 MBW68227.1 LNIX01000036 OXA39969.1 GBHO01030122 GBHO01030121 GBHO01030120 GBHO01030116 JAG13482.1 JAG13483.1 JAG13484.1 JAG13488.1
Proteomes
Pfam
Interpro
IPR008042
Retrotrans_Pao
+ More
IPR012337 RNaseH-like_sf
IPR001584 Integrase_cat-core
IPR005312 DUF1759
IPR041588 Integrase_H2C2
IPR008737 Peptidase_asp_put
IPR040676 DUF5641
IPR000477 RT_dom
IPR036397 RNaseH_sf
IPR021109 Peptidase_aspartic_dom_sf
IPR001969 Aspartic_peptidase_AS
IPR036875 Znf_CCHC_sf
IPR001878 Znf_CCHC
IPR041373 RT_RNaseH
IPR041577 RT_RNaseH_2
IPR001995 Peptidase_A2_cat
IPR013087 Znf_C2H2_type
IPR012337 RNaseH-like_sf
IPR001584 Integrase_cat-core
IPR005312 DUF1759
IPR041588 Integrase_H2C2
IPR008737 Peptidase_asp_put
IPR040676 DUF5641
IPR000477 RT_dom
IPR036397 RNaseH_sf
IPR021109 Peptidase_aspartic_dom_sf
IPR001969 Aspartic_peptidase_AS
IPR036875 Znf_CCHC_sf
IPR001878 Znf_CCHC
IPR041373 RT_RNaseH
IPR041577 RT_RNaseH_2
IPR001995 Peptidase_A2_cat
IPR013087 Znf_C2H2_type
Gene 3D
ProteinModelPortal
A0A3Q0JJJ7
A0A1S3DJJ0
A0A1S3DJ34
A0A1S3DJC0
A0A1Y1LIC8
A0A3Q0JFQ3
+ More
A0A3Q0JCL2 A0A1Y1LM33 A0A3Q0J7N1 A0A3Q0JBZ9 A0A3Q0J7K9 A0A3Q0J730 A0A1S3D744 A0A0J7KDN6 A0A1B6KIX6 X1XU04 J9JUK4 J9M7R2 A0A1B6MT91 X1WZF3 J9JL40 A0A0J7K7U0 X1WYQ7 A0A2S2NZ22 A0A0J7N890 A0A146KRI6 A0A1Y1LIG5 A0A1B6LLT3 A0A0A9WW84 A0A2S2P1X6 A0A1Y1KMX0 X1XU26 A0A2S2R2J0 A0A1W7R6A8 A0A1W7R6S6 A0A0A9ZHU0 A0A2S2QTX5 J9LPA9 A0A1U8N859 A0A087T9T6 A0A0J7N465 A0A2S2NHU0 X1X096 J9JLG6 J9JVZ7 A0A2H1X238 A0A1W7R6I7 A0A2S2PNZ4 A0A0N0PAG9 A0A226E780 A0A1W7R6N3 A0A226EPI5 A0A0A9WWW3 V9GZW9 J9LHB7 A0A2M4CW28 W8C1U1 A0A2M4CSC7 A0A226D417 A0A0A9X3X8
A0A3Q0JCL2 A0A1Y1LM33 A0A3Q0J7N1 A0A3Q0JBZ9 A0A3Q0J7K9 A0A3Q0J730 A0A1S3D744 A0A0J7KDN6 A0A1B6KIX6 X1XU04 J9JUK4 J9M7R2 A0A1B6MT91 X1WZF3 J9JL40 A0A0J7K7U0 X1WYQ7 A0A2S2NZ22 A0A0J7N890 A0A146KRI6 A0A1Y1LIG5 A0A1B6LLT3 A0A0A9WW84 A0A2S2P1X6 A0A1Y1KMX0 X1XU26 A0A2S2R2J0 A0A1W7R6A8 A0A1W7R6S6 A0A0A9ZHU0 A0A2S2QTX5 J9LPA9 A0A1U8N859 A0A087T9T6 A0A0J7N465 A0A2S2NHU0 X1X096 J9JLG6 J9JVZ7 A0A2H1X238 A0A1W7R6I7 A0A2S2PNZ4 A0A0N0PAG9 A0A226E780 A0A1W7R6N3 A0A226EPI5 A0A0A9WWW3 V9GZW9 J9LHB7 A0A2M4CW28 W8C1U1 A0A2M4CSC7 A0A226D417 A0A0A9X3X8
Ontologies
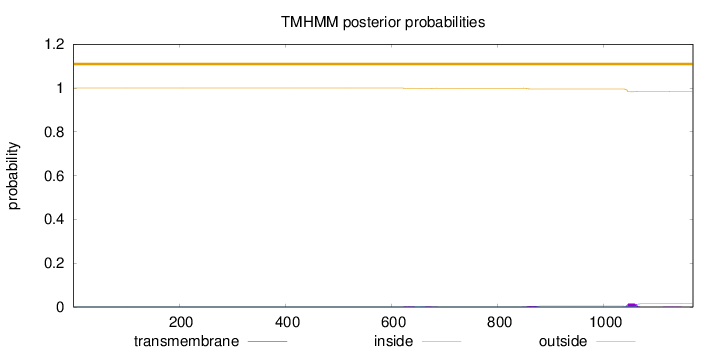
Topology
Length:
1168
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.47914
Exp number, first 60 AAs:
0.0004
Total prob of N-in:
0.00014
outside
1 - 1168
Population Genetic Test Statistics
Pi
0
Theta
0
Tajima's D
0
CLR
19.921586
CSRT
0
Interpretation
Uncertain
