Pre Gene Modal
BGIBMGA002208
Annotation
PREDICTED:_50S_ribosomal_protein_L1_[Papilio_xuthus]
Full name
E3 ubiquitin-protein ligase
Location in the cell
Mitochondrial Reliability : 2.156
Sequence
CDS
ATGGCTGGGACTATTTTCCGTTTGCTTTTGACCAATACCACTTCAGCCCACAAAAATATTACAACTATTAGATCCATAAACACAGGAACGGTATCATATGCAGCCAGGAAAGGTACAAGAGCTAAGGCACGTGCAAAAAAAATTCAAGTTGAAGTCACAAAAGTTGGTTTCATTCCGCACAATCAAAGAGGAAGAAAGGTAGTTAAAACAGTGGTGAACAAGCATTTCAGCGATGAGCACAAAGCGATACCCATTGACGATGTGTATCCTATGAAGTATTACAAATGGGTGGTTTACACAGCTGAGGATGCAGTCAAAGCCCACCAGGAGACCCACCACCCGACCATGTTTAATGCACCTGATGCCTTCATCTTTGCAAAAATTGAATTTAACATGACTGGTATAAAGCAGACTCGATTCATGGATAGTTTCACTCGTTTGTCCTTGCTGCCTCATCCTTTCCCAAGAGAAGAAGAGCGCACTATTCTTGCCTTCTGTAAAGGAGCAGAGTTAATCAAGGAATTACAAGATGCAGGTGCTACAGTGGCTGGAGGCACAGATATTGTTAAGAAGATACAGGATGGTCAGATCAAATTATTTGAATATGATTACGTTGTAGCCCATCCAAACATATTGACGGATTTAGTGCCAATCCGTGGTCTCATGAAGAAGCGGTTCCCTAACGTGAGGTCTGGAACCCTGGACCCTAATCTGAAGGAGTTGGTGAAAAAGTTTGCTGGAGGTATCCAATATAAAGTAATAAAAGACGAAGCGCAACCGAGTTTTGGATCTGTAGAAGTACCAGTTGGCCGGTTAAATATGGAACCAAAGCAAGTGGCTGAAAATATTGGAAAATTGCTAAAAGACGTGCAGGCTGTCCGCCCCAAGAGAGATGGGCTCTTCGTAACAAGATGCCTGTTGGTAAGTCCTCCATCTACGGAGCAATTAATGATTGATCCGTTCGTGTTCGTAGATAGAACACTCTCGAAAGAGGTTCAAGATGATAGCGATGACGAAGACGAGCCCGTGGCGGCCACAGCGTAA
Protein
MAGTIFRLLLTNTTSAHKNITTIRSINTGTVSYAARKGTRAKARAKKIQVEVTKVGFIPHNQRGRKVVKTVVNKHFSDEHKAIPIDDVYPMKYYKWVVYTAEDAVKAHQETHHPTMFNAPDAFIFAKIEFNMTGIKQTRFMDSFTRLSLLPHPFPREEERTILAFCKGAELIKELQDAGATVAGGTDIVKKIQDGQIKLFEYDYVVAHPNILTDLVPIRGLMKKRFPNVRSGTLDPNLKELVKKFAGGIQYKVIKDEAQPSFGSVEVPVGRLNMEPKQVAENIGKLLKDVQAVRPKRDGLFVTRCLLVSPPSTEQLMIDPFVFVDRTLSKEVQDDSDDEDEPVAATA
Summary
Description
E3 ubiquitin-protein ligase that mediates ubiquitination and subsequent proteasomal degradation of target proteins. E3 ubiquitin ligases accept ubiquitin from an E2 ubiquitin-conjugating enzyme in the form of a thioester and then directly transfers the ubiquitin to targeted substrates.
Catalytic Activity
S-ubiquitinyl-[E2 ubiquitin-conjugating enzyme]-L-cysteine + [acceptor protein]-L-lysine = [E2 ubiquitin-conjugating enzyme]-L-cysteine + N(6)-ubiquitinyl-[acceptor protein]-L-lysine.
Similarity
Belongs to the SINA (Seven in absentia) family.
Uniprot
H9IY75
A0A2A4JA26
A0A2H1WHT2
A0A194QHF4
A0A212EI69
A0A194REB1
+ More
A0A0L7LMS5 A0A1J1HWF5 A0A0M4EPU9 B4K7W0 B4JH28 A0A0K8TSC8 A0A1I8MEP7 B3NZU8 A0A1I8PVL7 A0A3B0JP31 J3JVE1 B4HLM6 B4M5Q3 N6UMU6 Q9VHT5 B4NAF2 A0A1W4VVT4 B3M0D1 B4PT35 A0A1A9UZ15 Q298H4 B4G3S1 A0A1A9Z9M8 A0A1B0GAV3 A0A0L0CRW0 A0A1B0BQ35 A0A336MRE8 A0A182JW95 T1GK75 A0A182LA50 A0A182RSX3 A0A1Q3FDU6 A0A0A1X437 A0A034VD95 A0A1Y9IW17 A0A182HLQ4 Q7PY42 A0A023EQC2 A0A1A9XG75 B0W376 Q17CA4 W8BXJ3 A0A182UEC9 A0A182UQU6 A0A084VZE8 A0A182LV61 A0A182H1U7 A0A1Y9HAI6 A0A182Y3U9 A0A182IP59 A0A1S4F875 A0A0K8WCC9 A0A1A9WY60 U5ET83 A0A182FPX3 A0A336MSJ3 A0A182PSG9 A0A2M4BS39 A0A2M4BSC4 W5JF00 A0A2M3Z8T8 A0A2M4AM80 A0A182NSV5 A0A182WSV1 A0A1L8DRT3 A0A1Y1LFT1 A0A1L8DRJ6 A0A1B6LGQ7 A0A0A9VZT1 A0A2P8XFF2 A0A067RP01 A0A069DSQ3 A0A1B6IVC6 R4WPM2 A0A1B6FXW7 A0A158NMT4 A0A195EXW9 D6X497 A0A2J7RS27 A0A1W4WEA6 A0A1W4WDB1 F4WTC0 A0A195BBY1 A0A3L8D4G1 A0A195C7M0 A0A1B6CAE8 A0A0P4VMZ6 A0A151WUS3 A0A0T6BA46 A0A151JM45 A0A088AET5 A0A1B0D1M8 E2ALJ5
A0A0L7LMS5 A0A1J1HWF5 A0A0M4EPU9 B4K7W0 B4JH28 A0A0K8TSC8 A0A1I8MEP7 B3NZU8 A0A1I8PVL7 A0A3B0JP31 J3JVE1 B4HLM6 B4M5Q3 N6UMU6 Q9VHT5 B4NAF2 A0A1W4VVT4 B3M0D1 B4PT35 A0A1A9UZ15 Q298H4 B4G3S1 A0A1A9Z9M8 A0A1B0GAV3 A0A0L0CRW0 A0A1B0BQ35 A0A336MRE8 A0A182JW95 T1GK75 A0A182LA50 A0A182RSX3 A0A1Q3FDU6 A0A0A1X437 A0A034VD95 A0A1Y9IW17 A0A182HLQ4 Q7PY42 A0A023EQC2 A0A1A9XG75 B0W376 Q17CA4 W8BXJ3 A0A182UEC9 A0A182UQU6 A0A084VZE8 A0A182LV61 A0A182H1U7 A0A1Y9HAI6 A0A182Y3U9 A0A182IP59 A0A1S4F875 A0A0K8WCC9 A0A1A9WY60 U5ET83 A0A182FPX3 A0A336MSJ3 A0A182PSG9 A0A2M4BS39 A0A2M4BSC4 W5JF00 A0A2M3Z8T8 A0A2M4AM80 A0A182NSV5 A0A182WSV1 A0A1L8DRT3 A0A1Y1LFT1 A0A1L8DRJ6 A0A1B6LGQ7 A0A0A9VZT1 A0A2P8XFF2 A0A067RP01 A0A069DSQ3 A0A1B6IVC6 R4WPM2 A0A1B6FXW7 A0A158NMT4 A0A195EXW9 D6X497 A0A2J7RS27 A0A1W4WEA6 A0A1W4WDB1 F4WTC0 A0A195BBY1 A0A3L8D4G1 A0A195C7M0 A0A1B6CAE8 A0A0P4VMZ6 A0A151WUS3 A0A0T6BA46 A0A151JM45 A0A088AET5 A0A1B0D1M8 E2ALJ5
EC Number
2.3.2.27
Pubmed
19121390
26354079
22118469
26227816
17994087
26369729
+ More
25315136 22516182 23537049 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 15632085 26108605 20966253 25830018 25348373 12364791 14747013 17210077 24945155 17510324 24495485 24438588 26483478 25244985 20920257 23761445 28004739 25401762 29403074 24845553 26334808 23691247 21347285 18362917 19820115 21719571 30249741 27129103 20798317
25315136 22516182 23537049 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 15632085 26108605 20966253 25830018 25348373 12364791 14747013 17210077 24945155 17510324 24495485 24438588 26483478 25244985 20920257 23761445 28004739 25401762 29403074 24845553 26334808 23691247 21347285 18362917 19820115 21719571 30249741 27129103 20798317
EMBL
BABH01038379
BABH01038380
BABH01038381
NWSH01002281
PCG68699.1
ODYU01008429
+ More
SOQ52024.1 KQ459011 KPJ04365.1 AGBW02014704 OWR41160.1 KQ460500 KPJ14266.1 JTDY01000596 KOB76526.1 CVRI01000026 CRK92349.1 CP012526 ALC46210.1 CH933806 EDW14294.1 CH916369 EDV92719.1 GDAI01000555 JAI17048.1 CH954181 EDV49946.1 OUUW01000007 SPP82673.1 BT127208 KB632192 AEE62170.1 ERL89871.1 CH480815 EDW43053.1 CH940652 EDW58979.1 APGK01017031 KB739995 ENN82021.1 AE014297 AY051800 AAF54214.2 AAK93224.1 CH964232 EDW80766.1 CH902617 EDV43137.1 CM000160 EDW96496.1 CM000070 EAL27981.1 CH479179 EDW24392.1 CCAG010010901 JRES01000007 KNC34927.1 JXJN01018364 UFQS01002153 UFQT01002153 SSX13352.1 SSX32786.1 CAQQ02392881 GFDL01009305 JAV25740.1 GBXI01008864 JAD05428.1 GAKP01017691 JAC41261.1 APCN01000910 AAAB01008987 EAA00834.3 GAPW01002328 JAC11270.1 DS231831 EDS31219.1 CH477310 EAT43971.1 GAMC01002553 JAC04003.1 ATLV01018748 KE525248 KFB43342.1 AXCM01000028 JXUM01023254 KQ560656 KXJ81406.1 AXCN02001149 GDHF01003799 JAI48515.1 GANO01002895 JAB56976.1 UFQT01001768 SSX31733.1 GGFJ01006691 MBW55832.1 GGFJ01006690 MBW55831.1 ADMH02001345 ETN62937.1 GGFM01004161 MBW24912.1 GGFK01008521 MBW41842.1 GFDF01005024 JAV09060.1 GEZM01056990 JAV72522.1 GFDF01005025 JAV09059.1 GEBQ01017095 JAT22882.1 GBHO01045494 JAF98109.1 PYGN01002348 PSN30702.1 KK852553 KDR21469.1 GBGD01002128 JAC86761.1 GECU01016810 JAS90896.1 AK417611 BAN20826.1 GECZ01014876 JAS54893.1 ADTU01020803 KQ981940 KYN32742.1 KQ971410 EEZ97527.1 NEVH01000595 PNF43636.1 GL888334 EGI62583.1 KQ976529 KYM81727.1 QOIP01000014 RLU15021.1 KQ978231 KYM96161.1 GEDC01026979 GEDC01019601 GEDC01010631 JAS10319.1 JAS17697.1 JAS26667.1 GDKW01002884 JAI53711.1 KQ982729 KYQ51578.1 LJIG01002739 KRT84201.1 KQ978946 KYN27399.1 AJVK01022206 GL440609 EFN65697.1
SOQ52024.1 KQ459011 KPJ04365.1 AGBW02014704 OWR41160.1 KQ460500 KPJ14266.1 JTDY01000596 KOB76526.1 CVRI01000026 CRK92349.1 CP012526 ALC46210.1 CH933806 EDW14294.1 CH916369 EDV92719.1 GDAI01000555 JAI17048.1 CH954181 EDV49946.1 OUUW01000007 SPP82673.1 BT127208 KB632192 AEE62170.1 ERL89871.1 CH480815 EDW43053.1 CH940652 EDW58979.1 APGK01017031 KB739995 ENN82021.1 AE014297 AY051800 AAF54214.2 AAK93224.1 CH964232 EDW80766.1 CH902617 EDV43137.1 CM000160 EDW96496.1 CM000070 EAL27981.1 CH479179 EDW24392.1 CCAG010010901 JRES01000007 KNC34927.1 JXJN01018364 UFQS01002153 UFQT01002153 SSX13352.1 SSX32786.1 CAQQ02392881 GFDL01009305 JAV25740.1 GBXI01008864 JAD05428.1 GAKP01017691 JAC41261.1 APCN01000910 AAAB01008987 EAA00834.3 GAPW01002328 JAC11270.1 DS231831 EDS31219.1 CH477310 EAT43971.1 GAMC01002553 JAC04003.1 ATLV01018748 KE525248 KFB43342.1 AXCM01000028 JXUM01023254 KQ560656 KXJ81406.1 AXCN02001149 GDHF01003799 JAI48515.1 GANO01002895 JAB56976.1 UFQT01001768 SSX31733.1 GGFJ01006691 MBW55832.1 GGFJ01006690 MBW55831.1 ADMH02001345 ETN62937.1 GGFM01004161 MBW24912.1 GGFK01008521 MBW41842.1 GFDF01005024 JAV09060.1 GEZM01056990 JAV72522.1 GFDF01005025 JAV09059.1 GEBQ01017095 JAT22882.1 GBHO01045494 JAF98109.1 PYGN01002348 PSN30702.1 KK852553 KDR21469.1 GBGD01002128 JAC86761.1 GECU01016810 JAS90896.1 AK417611 BAN20826.1 GECZ01014876 JAS54893.1 ADTU01020803 KQ981940 KYN32742.1 KQ971410 EEZ97527.1 NEVH01000595 PNF43636.1 GL888334 EGI62583.1 KQ976529 KYM81727.1 QOIP01000014 RLU15021.1 KQ978231 KYM96161.1 GEDC01026979 GEDC01019601 GEDC01010631 JAS10319.1 JAS17697.1 JAS26667.1 GDKW01002884 JAI53711.1 KQ982729 KYQ51578.1 LJIG01002739 KRT84201.1 KQ978946 KYN27399.1 AJVK01022206 GL440609 EFN65697.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000053240
UP000037510
+ More
UP000183832 UP000092553 UP000009192 UP000001070 UP000095301 UP000008711 UP000095300 UP000268350 UP000030742 UP000001292 UP000008792 UP000019118 UP000000803 UP000007798 UP000192221 UP000007801 UP000002282 UP000078200 UP000001819 UP000008744 UP000092445 UP000092444 UP000037069 UP000092460 UP000075881 UP000015102 UP000075882 UP000075900 UP000075920 UP000075840 UP000007062 UP000092443 UP000002320 UP000008820 UP000075902 UP000075903 UP000030765 UP000075883 UP000069940 UP000249989 UP000075886 UP000076408 UP000075880 UP000091820 UP000069272 UP000075885 UP000000673 UP000075884 UP000076407 UP000245037 UP000027135 UP000005205 UP000078541 UP000007266 UP000235965 UP000192223 UP000007755 UP000078540 UP000279307 UP000078542 UP000075809 UP000078492 UP000005203 UP000092462 UP000000311
UP000183832 UP000092553 UP000009192 UP000001070 UP000095301 UP000008711 UP000095300 UP000268350 UP000030742 UP000001292 UP000008792 UP000019118 UP000000803 UP000007798 UP000192221 UP000007801 UP000002282 UP000078200 UP000001819 UP000008744 UP000092445 UP000092444 UP000037069 UP000092460 UP000075881 UP000015102 UP000075882 UP000075900 UP000075920 UP000075840 UP000007062 UP000092443 UP000002320 UP000008820 UP000075902 UP000075903 UP000030765 UP000075883 UP000069940 UP000249989 UP000075886 UP000076408 UP000075880 UP000091820 UP000069272 UP000075885 UP000000673 UP000075884 UP000076407 UP000245037 UP000027135 UP000005205 UP000078541 UP000007266 UP000235965 UP000192223 UP000007755 UP000078540 UP000279307 UP000078542 UP000075809 UP000078492 UP000005203 UP000092462 UP000000311
Interpro
Gene 3D
ProteinModelPortal
H9IY75
A0A2A4JA26
A0A2H1WHT2
A0A194QHF4
A0A212EI69
A0A194REB1
+ More
A0A0L7LMS5 A0A1J1HWF5 A0A0M4EPU9 B4K7W0 B4JH28 A0A0K8TSC8 A0A1I8MEP7 B3NZU8 A0A1I8PVL7 A0A3B0JP31 J3JVE1 B4HLM6 B4M5Q3 N6UMU6 Q9VHT5 B4NAF2 A0A1W4VVT4 B3M0D1 B4PT35 A0A1A9UZ15 Q298H4 B4G3S1 A0A1A9Z9M8 A0A1B0GAV3 A0A0L0CRW0 A0A1B0BQ35 A0A336MRE8 A0A182JW95 T1GK75 A0A182LA50 A0A182RSX3 A0A1Q3FDU6 A0A0A1X437 A0A034VD95 A0A1Y9IW17 A0A182HLQ4 Q7PY42 A0A023EQC2 A0A1A9XG75 B0W376 Q17CA4 W8BXJ3 A0A182UEC9 A0A182UQU6 A0A084VZE8 A0A182LV61 A0A182H1U7 A0A1Y9HAI6 A0A182Y3U9 A0A182IP59 A0A1S4F875 A0A0K8WCC9 A0A1A9WY60 U5ET83 A0A182FPX3 A0A336MSJ3 A0A182PSG9 A0A2M4BS39 A0A2M4BSC4 W5JF00 A0A2M3Z8T8 A0A2M4AM80 A0A182NSV5 A0A182WSV1 A0A1L8DRT3 A0A1Y1LFT1 A0A1L8DRJ6 A0A1B6LGQ7 A0A0A9VZT1 A0A2P8XFF2 A0A067RP01 A0A069DSQ3 A0A1B6IVC6 R4WPM2 A0A1B6FXW7 A0A158NMT4 A0A195EXW9 D6X497 A0A2J7RS27 A0A1W4WEA6 A0A1W4WDB1 F4WTC0 A0A195BBY1 A0A3L8D4G1 A0A195C7M0 A0A1B6CAE8 A0A0P4VMZ6 A0A151WUS3 A0A0T6BA46 A0A151JM45 A0A088AET5 A0A1B0D1M8 E2ALJ5
A0A0L7LMS5 A0A1J1HWF5 A0A0M4EPU9 B4K7W0 B4JH28 A0A0K8TSC8 A0A1I8MEP7 B3NZU8 A0A1I8PVL7 A0A3B0JP31 J3JVE1 B4HLM6 B4M5Q3 N6UMU6 Q9VHT5 B4NAF2 A0A1W4VVT4 B3M0D1 B4PT35 A0A1A9UZ15 Q298H4 B4G3S1 A0A1A9Z9M8 A0A1B0GAV3 A0A0L0CRW0 A0A1B0BQ35 A0A336MRE8 A0A182JW95 T1GK75 A0A182LA50 A0A182RSX3 A0A1Q3FDU6 A0A0A1X437 A0A034VD95 A0A1Y9IW17 A0A182HLQ4 Q7PY42 A0A023EQC2 A0A1A9XG75 B0W376 Q17CA4 W8BXJ3 A0A182UEC9 A0A182UQU6 A0A084VZE8 A0A182LV61 A0A182H1U7 A0A1Y9HAI6 A0A182Y3U9 A0A182IP59 A0A1S4F875 A0A0K8WCC9 A0A1A9WY60 U5ET83 A0A182FPX3 A0A336MSJ3 A0A182PSG9 A0A2M4BS39 A0A2M4BSC4 W5JF00 A0A2M3Z8T8 A0A2M4AM80 A0A182NSV5 A0A182WSV1 A0A1L8DRT3 A0A1Y1LFT1 A0A1L8DRJ6 A0A1B6LGQ7 A0A0A9VZT1 A0A2P8XFF2 A0A067RP01 A0A069DSQ3 A0A1B6IVC6 R4WPM2 A0A1B6FXW7 A0A158NMT4 A0A195EXW9 D6X497 A0A2J7RS27 A0A1W4WEA6 A0A1W4WDB1 F4WTC0 A0A195BBY1 A0A3L8D4G1 A0A195C7M0 A0A1B6CAE8 A0A0P4VMZ6 A0A151WUS3 A0A0T6BA46 A0A151JM45 A0A088AET5 A0A1B0D1M8 E2ALJ5
PDB
2FTC
E-value=2.81262e-15,
Score=199
Ontologies
GO
PANTHER
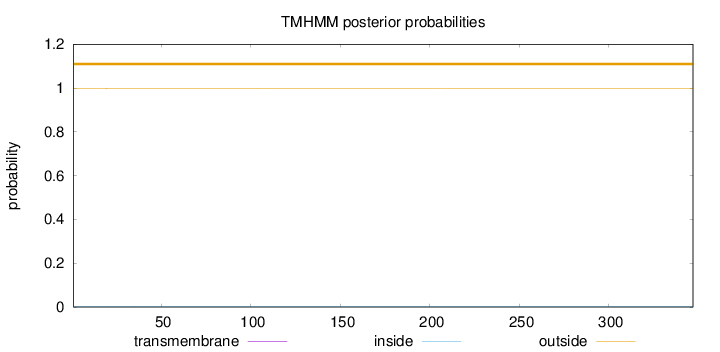
Topology
Length:
347
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00365
Exp number, first 60 AAs:
0.00114
Total prob of N-in:
0.00265
outside
1 - 347
Population Genetic Test Statistics
Pi
5.042268
Theta
17.3216
Tajima's D
-1.262076
CLR
17.920576
CSRT
0.0902454877256137
Interpretation
Uncertain
