Gene
KWMTBOMO03665
Pre Gene Modal
BGIBMGA002224
Annotation
PREDICTED:_uncharacterized_protein_LOC105383792_[Plutella_xylostella]
Location in the cell
PlasmaMembrane Reliability : 1.288
Sequence
CDS
ATGGAGAAAATAAACTGCAGTTTGTTAGCAATATCGCTTTGTGCATTGTTGTGGGGTGTGGCCGGTCAGTACGAATGGCAAATACGCGATGCTTTTGACGAAGTCCGAGGGAAGATGGACAAGATTAATAGCGAAAATTGTTACATTAGTCATCTCGATGACTTGTACCTCTCCGAAGATTCTGTTTCTCACCATCCCGACGTGAAAGAGATCAACATCAACCCGGTATTCCCAAACCGTACCGCGATGCTGCATTTGCACAATATGGCCATGAATAGAGCTTTTTTCTGGAGTTTCGTTCTCCAAACCAGATTTATTAGACCCGCCATCAATGATACCTACGATCCTGGTATGATGTATTATTTCCTTTCGGCCGTTGCCGATGTGGCTGCCAATCCCTACATAAACGCTAGTGCAATATATTTTTCCCCAAATATGTCGTATACCTCGTCGTACAGAGGGTTCTTTAACAAGACCCTGCCGCGTTTTGCTCCCAGAGCATTCCGAGCTGATGATTTTAACGATCCTGTACACTTACAAAAAATATCTACTATGAACACTTTTATAATTGAAGATTTAGGTGCTTTCGAACCAGATAGCTTATCGAAAGACTATACGTCAGAATTCTATAGGACTAATGAGTGGTATAAGGTGTGGCTACCTGACAGAGTGGAAAGGCGTCATGATACTAAGACTACATATCAAGTAGAGATCAGATATGCTAACAATACAAACGAAACATTCACATTTCATGGTCCACCAGGGAATGATGAGACTCCTGGTCCAGTGAACTGGACCCGTCCATACTTTGATTGCGGTCGGCTCAGCAAGTGGTTAGTGGGTGCGGTTTCGCCTGTTGCTGATATATACCCTCGACACACGCAATTCAGACATATTGAATACCCCACTTACACGGCCGCCGTCGTTATGGAAATGGATTATGAGAGGATCGACATAAACCAGTGTCCTCCGAGTCCGGGCAACGACCGACCGAACAAGTTCGCATCGACGGCTCGATGCAAAGAAACAACAGAGTGCGAGCCGTTGGACGGGTGGGGCTTCCGTCGCGGCGGCTACCAGTGTCGCTGCCGGCCCGGGTACCGGCTGCCCAGCATCGTGCGCCGCCCCTACCTGGGGGAGATCATCGAGCGGGCCACCTCCGACCAGTACTACAACAACTTCGACTGCTCCAAGATTGGATGGATCCAGGGCCTGCCGTCGGAGCTGGAGAAGGCGCCGGCGTTCCTGCGCGCGGTGTACCGGGACCGCTACCACGAGTACGTGCAGGCGGCCGCCGGGCCCGCCGCGCTGCACGCGCCGCGACCCAACGTCTACGAGATGCTCAACTTCATCCGCGGCGTGCGCCCGGACAACTGCTCCCGGTACAACCCCAGCGACCTGGTCCTGAACGGTGATTTGGCGTTCGGAGTCGAGAAGCAGCTGGAGAACCAAGCCAAGATGGCTCTGAGGCTGGCCAACTTCATCAGCGCGTTCTTGCAGGTGTCGGATCCACAAGAAGTGTTTAGCGGGAACCGAGTCGCCGACAAACCCCTGACCGAAGACCAGATGATAGGCGAGACCCTGGCCATCGTGCTCGGAGACTCCAAGATCTGGTCCGCCGGCACGTACTGGGACCGAGAGAAGTTCCCGAACCGCACGTACTTCGCGCCCTTCGCCTACAAGACCGAGCTCAACGTGAGGAAGTTCCGTCTCGAGGACCTGGCGCGACTCAACTCCACAGAGGAAGTGTACACGAACAAGCCCTGGTTCCAGTTCCTGAAGCAGCGCTGGTCGAGCAACTTCGACAGCCTCGAGCAATACTTCCTCAAGATGAACATCCGCGACTCGGAGGCCGGCAAGTACCTCAAGCACTACGAGCGCTTCCCGCACTGGTACCGGGCGGCGGCGCTGCGCCACGGACACTGGACGCAGCCCTACTACGACTGCGGGGGCCGCGTGCCGCAGTGGGCCGTCACCTACGCCGCGCCCTTCTTCGGCTGGGACAGCGTCAAAGTCAAACTCGAGTTCAAAGGTGCGGTGGCGGTGACGATGTCATTGCTGTCTCTGGACATCAACCAGTGCCCGGACAAGTACTACGTGTCCAACGCATTCAAGGGAACGGATAAATGCGACAGGAGCTCGTCTTATTGCGTCCCGATCCTGGGCCGCGGCTACGAGGCGGGCGGGTACAAGTGCGAGTGCCTGCAGGGCTTCGAGTACCCGTTCGAGGACCCCACCACGTACTACGACGGGCAGATCGTGGAGGCCGAGTTCCAGAACATCATACTCGACAAGGAGACGCGCATCGACATGTTTAAGTGCCGCCTGGCCGGCGCCGCAGCCCTGGGGCTCAGCCCCGCCCTGCTGCTCGCGCTGGTAGCCACGCTGGTCGGCCTGCGGTAG
Protein
MEKINCSLLAISLCALLWGVAGQYEWQIRDAFDEVRGKMDKINSENCYISHLDDLYLSEDSVSHHPDVKEININPVFPNRTAMLHLHNMAMNRAFFWSFVLQTRFIRPAINDTYDPGMMYYFLSAVADVAANPYINASAIYFSPNMSYTSSYRGFFNKTLPRFAPRAFRADDFNDPVHLQKISTMNTFIIEDLGAFEPDSLSKDYTSEFYRTNEWYKVWLPDRVERRHDTKTTYQVEIRYANNTNETFTFHGPPGNDETPGPVNWTRPYFDCGRLSKWLVGAVSPVADIYPRHTQFRHIEYPTYTAAVVMEMDYERIDINQCPPSPGNDRPNKFASTARCKETTECEPLDGWGFRRGGYQCRCRPGYRLPSIVRRPYLGEIIERATSDQYYNNFDCSKIGWIQGLPSELEKAPAFLRAVYRDRYHEYVQAAAGPAALHAPRPNVYEMLNFIRGVRPDNCSRYNPSDLVLNGDLAFGVEKQLENQAKMALRLANFISAFLQVSDPQEVFSGNRVADKPLTEDQMIGETLAIVLGDSKIWSAGTYWDREKFPNRTYFAPFAYKTELNVRKFRLEDLARLNSTEEVYTNKPWFQFLKQRWSSNFDSLEQYFLKMNIRDSEAGKYLKHYERFPHWYRAAALRHGHWTQPYYDCGGRVPQWAVTYAAPFFGWDSVKVKLEFKGAVAVTMSLLSLDINQCPDKYYVSNAFKGTDKCDRSSSYCVPILGRGYEAGGYKCECLQGFEYPFEDPTTYYDGQIVEAEFQNIILDKETRIDMFKCRLAGAAALGLSPALLLALVATLVGLR
Summary
Uniprot
A0A2A4JAI0
A0A194R9F8
A0A194QLP5
A0A212EI47
A0A2H1WG35
A0A067RR18
+ More
U5ENY5 A0A1A9VEG2 A0A026VWX1 A0A1A9WY97 A0A1A9Z8C0 A0A1B0BBB9 A0A1I8NMT1 A0A1B0FF18 A0A0C9QD93 A0A1I8MS57 A0A195CS51 E2AQJ5 A0A151HYW2 A0A158NSH1 A0A1A9XNC1 A0A1L8DKF3 A0A0K8V3R9 A0A0A1XEX2 A0A034WBA7 A0A195FT86 A0A0L7RDL4 A0A0M8ZYJ6 W8B0V8 A0A1B6GGM7 A0A151J4B0 A0A1B6IF03 B4LWK7 B4K8W0 A0A2A3EFN2 B4NFT4 A0A1W4UXF3 A0A3B0JQ97 B4GZE9 A0A0M4F5B3 Q29AX1 A0A310SL49 B4II78 E0VKM8 A0A0A9W679 B4R1S4 Q8IN49 B3P331 A0A336KV44 B4JUD6 B3LZV3 A0A2M3YYZ6 B4PPS4 A0A2M4A6Y3 A0A182FP27 A0A1Y1KSV5 D6WCC0 A0A1J1I4F5 A0A2M4BE39 A0A2M4BED6 E2B838 W5JIL3 A0A182V879 A0A182X9V8 A0A182KA74 A0A182TUM4 A0A182L953 Q7PXI4 A0A182HKU1 A0A182M7C6 A0A336KWP4 A0A182RPL7 N6SX55 A0A182GZA6 A0A182Y579 A0A182N5F8 A0A182QTM0 A0A182P2E6 Q17GV4 A0A084VW52 A0A182VRB0 B0W9C9 A0A182IWV1 A0A336M0I1 A0A0P4VPW0 A0A088AN00 T1HTV6 E9HJM8 A0A0P6H196 A0A162TCC1 A0A0P5DDS5 A0A0P5ZCQ0 K7J030 A0A0P5Q9A7 A0A0P5TM39 A0A0P5VQV4 A0A0P6BYQ9 A0A1B6D655 A0A226DYF3 A0A1B0DPV8
U5ENY5 A0A1A9VEG2 A0A026VWX1 A0A1A9WY97 A0A1A9Z8C0 A0A1B0BBB9 A0A1I8NMT1 A0A1B0FF18 A0A0C9QD93 A0A1I8MS57 A0A195CS51 E2AQJ5 A0A151HYW2 A0A158NSH1 A0A1A9XNC1 A0A1L8DKF3 A0A0K8V3R9 A0A0A1XEX2 A0A034WBA7 A0A195FT86 A0A0L7RDL4 A0A0M8ZYJ6 W8B0V8 A0A1B6GGM7 A0A151J4B0 A0A1B6IF03 B4LWK7 B4K8W0 A0A2A3EFN2 B4NFT4 A0A1W4UXF3 A0A3B0JQ97 B4GZE9 A0A0M4F5B3 Q29AX1 A0A310SL49 B4II78 E0VKM8 A0A0A9W679 B4R1S4 Q8IN49 B3P331 A0A336KV44 B4JUD6 B3LZV3 A0A2M3YYZ6 B4PPS4 A0A2M4A6Y3 A0A182FP27 A0A1Y1KSV5 D6WCC0 A0A1J1I4F5 A0A2M4BE39 A0A2M4BED6 E2B838 W5JIL3 A0A182V879 A0A182X9V8 A0A182KA74 A0A182TUM4 A0A182L953 Q7PXI4 A0A182HKU1 A0A182M7C6 A0A336KWP4 A0A182RPL7 N6SX55 A0A182GZA6 A0A182Y579 A0A182N5F8 A0A182QTM0 A0A182P2E6 Q17GV4 A0A084VW52 A0A182VRB0 B0W9C9 A0A182IWV1 A0A336M0I1 A0A0P4VPW0 A0A088AN00 T1HTV6 E9HJM8 A0A0P6H196 A0A162TCC1 A0A0P5DDS5 A0A0P5ZCQ0 K7J030 A0A0P5Q9A7 A0A0P5TM39 A0A0P5VQV4 A0A0P6BYQ9 A0A1B6D655 A0A226DYF3 A0A1B0DPV8
Pubmed
26354079
22118469
24845553
24508170
30249741
25315136
+ More
20798317 21347285 25830018 25348373 24495485 17994087 15632085 20566863 25401762 26823975 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 28004739 18362917 19820115 20920257 23761445 20966253 12364791 14747013 17210077 23537049 26483478 25244985 17510324 24438588 27129103 21292972 20075255
20798317 21347285 25830018 25348373 24495485 17994087 15632085 20566863 25401762 26823975 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 28004739 18362917 19820115 20920257 23761445 20966253 12364791 14747013 17210077 23537049 26483478 25244985 17510324 24438588 27129103 21292972 20075255
EMBL
NWSH01002281
PCG68698.1
KQ460500
KPJ14267.1
KQ459011
KPJ04366.1
+ More
AGBW02014704 OWR41159.1 ODYU01008429 SOQ52021.1 KK852517 KDR22174.1 GANO01004947 JAB54924.1 KK107652 QOIP01000003 EZA48293.1 RLU25175.1 JXJN01011388 CCAG010009824 GBYB01012363 JAG82130.1 KQ977329 KYN03563.1 GL441773 EFN64304.1 KQ976719 KYM76760.1 ADTU01024910 GFDF01007260 JAV06824.1 GDHF01018747 JAI33567.1 GBXI01004353 JAD09939.1 GAKP01007884 JAC51068.1 KQ981276 KYN43517.1 KQ414613 KOC68943.1 KQ435824 KOX72199.1 GAMC01014433 GAMC01014431 JAB92122.1 GECZ01008177 JAS61592.1 KQ980159 KYN17422.1 GECU01022233 JAS85473.1 CH940650 EDW67672.1 CH933806 EDW16557.1 KZ288269 PBC30019.1 CH964251 EDW83151.1 OUUW01000007 SPP83093.1 CH479198 EDW28167.1 CP012526 ALC46707.1 CM000070 EAL27228.1 KQ761768 OAD57109.1 CH480842 EDW49622.1 DS235250 EEB13934.1 GBHO01040693 GBHO01040691 GBHO01016503 GBHO01016492 GBRD01016303 GDHC01006438 JAG02911.1 JAG02913.1 JAG27101.1 JAG27112.1 JAG49523.1 JAQ12191.1 CM000364 EDX12183.1 AE014297 BT083386 AAN13829.2 ACQ66047.1 CH954181 EDV48345.1 UFQS01000993 UFQT01000993 SSX08322.1 SSX28345.1 CH916374 EDV91106.1 CH902617 EDV43097.1 GGFM01000749 MBW21500.1 CM000160 EDW96174.1 GGFK01003236 MBW36557.1 GEZM01080993 JAV61947.1 KQ971316 EEZ98791.1 CVRI01000039 CRK94642.1 GGFJ01002179 MBW51320.1 GGFJ01002180 MBW51321.1 GL446282 EFN88130.1 ADMH02001372 ETN62735.1 AAAB01008987 EAA01581.4 APCN01000869 AXCM01010104 SSX08319.1 SSX28342.1 APGK01053370 APGK01053371 APGK01053372 KB741223 ENN72369.1 JXUM01099340 JXUM01099341 KQ564484 KXJ72213.1 AXCN02001173 CH477255 EAT45901.1 ATLV01017444 KE525168 KFB42196.1 DS231863 EDS39989.1 UFQT01000380 SSX23745.1 GDKW01001554 JAI55041.1 ACPB03009759 GL732663 EFX68045.1 GDIQ01038253 JAN56484.1 LRGB01000005 KZS22076.1 GDIP01173811 GDIP01118628 JAJ49591.1 GDIP01058340 JAM45375.1 GDIQ01117773 JAL33953.1 GDIP01125801 JAL77913.1 GDIP01096691 JAM07024.1 GDIP01023268 JAM80447.1 GEDC01016145 JAS21153.1 LNIX01000009 OXA50259.1 AJVK01018633 AJVK01018634
AGBW02014704 OWR41159.1 ODYU01008429 SOQ52021.1 KK852517 KDR22174.1 GANO01004947 JAB54924.1 KK107652 QOIP01000003 EZA48293.1 RLU25175.1 JXJN01011388 CCAG010009824 GBYB01012363 JAG82130.1 KQ977329 KYN03563.1 GL441773 EFN64304.1 KQ976719 KYM76760.1 ADTU01024910 GFDF01007260 JAV06824.1 GDHF01018747 JAI33567.1 GBXI01004353 JAD09939.1 GAKP01007884 JAC51068.1 KQ981276 KYN43517.1 KQ414613 KOC68943.1 KQ435824 KOX72199.1 GAMC01014433 GAMC01014431 JAB92122.1 GECZ01008177 JAS61592.1 KQ980159 KYN17422.1 GECU01022233 JAS85473.1 CH940650 EDW67672.1 CH933806 EDW16557.1 KZ288269 PBC30019.1 CH964251 EDW83151.1 OUUW01000007 SPP83093.1 CH479198 EDW28167.1 CP012526 ALC46707.1 CM000070 EAL27228.1 KQ761768 OAD57109.1 CH480842 EDW49622.1 DS235250 EEB13934.1 GBHO01040693 GBHO01040691 GBHO01016503 GBHO01016492 GBRD01016303 GDHC01006438 JAG02911.1 JAG02913.1 JAG27101.1 JAG27112.1 JAG49523.1 JAQ12191.1 CM000364 EDX12183.1 AE014297 BT083386 AAN13829.2 ACQ66047.1 CH954181 EDV48345.1 UFQS01000993 UFQT01000993 SSX08322.1 SSX28345.1 CH916374 EDV91106.1 CH902617 EDV43097.1 GGFM01000749 MBW21500.1 CM000160 EDW96174.1 GGFK01003236 MBW36557.1 GEZM01080993 JAV61947.1 KQ971316 EEZ98791.1 CVRI01000039 CRK94642.1 GGFJ01002179 MBW51320.1 GGFJ01002180 MBW51321.1 GL446282 EFN88130.1 ADMH02001372 ETN62735.1 AAAB01008987 EAA01581.4 APCN01000869 AXCM01010104 SSX08319.1 SSX28342.1 APGK01053370 APGK01053371 APGK01053372 KB741223 ENN72369.1 JXUM01099340 JXUM01099341 KQ564484 KXJ72213.1 AXCN02001173 CH477255 EAT45901.1 ATLV01017444 KE525168 KFB42196.1 DS231863 EDS39989.1 UFQT01000380 SSX23745.1 GDKW01001554 JAI55041.1 ACPB03009759 GL732663 EFX68045.1 GDIQ01038253 JAN56484.1 LRGB01000005 KZS22076.1 GDIP01173811 GDIP01118628 JAJ49591.1 GDIP01058340 JAM45375.1 GDIQ01117773 JAL33953.1 GDIP01125801 JAL77913.1 GDIP01096691 JAM07024.1 GDIP01023268 JAM80447.1 GEDC01016145 JAS21153.1 LNIX01000009 OXA50259.1 AJVK01018633 AJVK01018634
Proteomes
UP000218220
UP000053240
UP000053268
UP000007151
UP000027135
UP000078200
+ More
UP000053097 UP000279307 UP000091820 UP000092445 UP000092460 UP000095300 UP000092444 UP000095301 UP000078542 UP000000311 UP000078540 UP000005205 UP000092443 UP000078541 UP000053825 UP000053105 UP000078492 UP000008792 UP000009192 UP000242457 UP000007798 UP000192221 UP000268350 UP000008744 UP000092553 UP000001819 UP000001292 UP000009046 UP000000304 UP000000803 UP000008711 UP000001070 UP000007801 UP000002282 UP000069272 UP000007266 UP000183832 UP000008237 UP000000673 UP000075903 UP000076407 UP000075881 UP000075902 UP000075882 UP000007062 UP000075840 UP000075883 UP000075900 UP000019118 UP000069940 UP000249989 UP000076408 UP000075884 UP000075886 UP000075885 UP000008820 UP000030765 UP000075920 UP000002320 UP000075880 UP000005203 UP000015103 UP000000305 UP000076858 UP000002358 UP000198287 UP000092462
UP000053097 UP000279307 UP000091820 UP000092445 UP000092460 UP000095300 UP000092444 UP000095301 UP000078542 UP000000311 UP000078540 UP000005205 UP000092443 UP000078541 UP000053825 UP000053105 UP000078492 UP000008792 UP000009192 UP000242457 UP000007798 UP000192221 UP000268350 UP000008744 UP000092553 UP000001819 UP000001292 UP000009046 UP000000304 UP000000803 UP000008711 UP000001070 UP000007801 UP000002282 UP000069272 UP000007266 UP000183832 UP000008237 UP000000673 UP000075903 UP000076407 UP000075881 UP000075902 UP000075882 UP000007062 UP000075840 UP000075883 UP000075900 UP000019118 UP000069940 UP000249989 UP000076408 UP000075884 UP000075886 UP000075885 UP000008820 UP000030765 UP000075920 UP000002320 UP000075880 UP000005203 UP000015103 UP000000305 UP000076858 UP000002358 UP000198287 UP000092462
PRIDE
Pfam
PF02487 CLN3
SUPFAM
SSF103473
SSF103473
CDD
ProteinModelPortal
A0A2A4JAI0
A0A194R9F8
A0A194QLP5
A0A212EI47
A0A2H1WG35
A0A067RR18
+ More
U5ENY5 A0A1A9VEG2 A0A026VWX1 A0A1A9WY97 A0A1A9Z8C0 A0A1B0BBB9 A0A1I8NMT1 A0A1B0FF18 A0A0C9QD93 A0A1I8MS57 A0A195CS51 E2AQJ5 A0A151HYW2 A0A158NSH1 A0A1A9XNC1 A0A1L8DKF3 A0A0K8V3R9 A0A0A1XEX2 A0A034WBA7 A0A195FT86 A0A0L7RDL4 A0A0M8ZYJ6 W8B0V8 A0A1B6GGM7 A0A151J4B0 A0A1B6IF03 B4LWK7 B4K8W0 A0A2A3EFN2 B4NFT4 A0A1W4UXF3 A0A3B0JQ97 B4GZE9 A0A0M4F5B3 Q29AX1 A0A310SL49 B4II78 E0VKM8 A0A0A9W679 B4R1S4 Q8IN49 B3P331 A0A336KV44 B4JUD6 B3LZV3 A0A2M3YYZ6 B4PPS4 A0A2M4A6Y3 A0A182FP27 A0A1Y1KSV5 D6WCC0 A0A1J1I4F5 A0A2M4BE39 A0A2M4BED6 E2B838 W5JIL3 A0A182V879 A0A182X9V8 A0A182KA74 A0A182TUM4 A0A182L953 Q7PXI4 A0A182HKU1 A0A182M7C6 A0A336KWP4 A0A182RPL7 N6SX55 A0A182GZA6 A0A182Y579 A0A182N5F8 A0A182QTM0 A0A182P2E6 Q17GV4 A0A084VW52 A0A182VRB0 B0W9C9 A0A182IWV1 A0A336M0I1 A0A0P4VPW0 A0A088AN00 T1HTV6 E9HJM8 A0A0P6H196 A0A162TCC1 A0A0P5DDS5 A0A0P5ZCQ0 K7J030 A0A0P5Q9A7 A0A0P5TM39 A0A0P5VQV4 A0A0P6BYQ9 A0A1B6D655 A0A226DYF3 A0A1B0DPV8
U5ENY5 A0A1A9VEG2 A0A026VWX1 A0A1A9WY97 A0A1A9Z8C0 A0A1B0BBB9 A0A1I8NMT1 A0A1B0FF18 A0A0C9QD93 A0A1I8MS57 A0A195CS51 E2AQJ5 A0A151HYW2 A0A158NSH1 A0A1A9XNC1 A0A1L8DKF3 A0A0K8V3R9 A0A0A1XEX2 A0A034WBA7 A0A195FT86 A0A0L7RDL4 A0A0M8ZYJ6 W8B0V8 A0A1B6GGM7 A0A151J4B0 A0A1B6IF03 B4LWK7 B4K8W0 A0A2A3EFN2 B4NFT4 A0A1W4UXF3 A0A3B0JQ97 B4GZE9 A0A0M4F5B3 Q29AX1 A0A310SL49 B4II78 E0VKM8 A0A0A9W679 B4R1S4 Q8IN49 B3P331 A0A336KV44 B4JUD6 B3LZV3 A0A2M3YYZ6 B4PPS4 A0A2M4A6Y3 A0A182FP27 A0A1Y1KSV5 D6WCC0 A0A1J1I4F5 A0A2M4BE39 A0A2M4BED6 E2B838 W5JIL3 A0A182V879 A0A182X9V8 A0A182KA74 A0A182TUM4 A0A182L953 Q7PXI4 A0A182HKU1 A0A182M7C6 A0A336KWP4 A0A182RPL7 N6SX55 A0A182GZA6 A0A182Y579 A0A182N5F8 A0A182QTM0 A0A182P2E6 Q17GV4 A0A084VW52 A0A182VRB0 B0W9C9 A0A182IWV1 A0A336M0I1 A0A0P4VPW0 A0A088AN00 T1HTV6 E9HJM8 A0A0P6H196 A0A162TCC1 A0A0P5DDS5 A0A0P5ZCQ0 K7J030 A0A0P5Q9A7 A0A0P5TM39 A0A0P5VQV4 A0A0P6BYQ9 A0A1B6D655 A0A226DYF3 A0A1B0DPV8
Ontologies
Topology
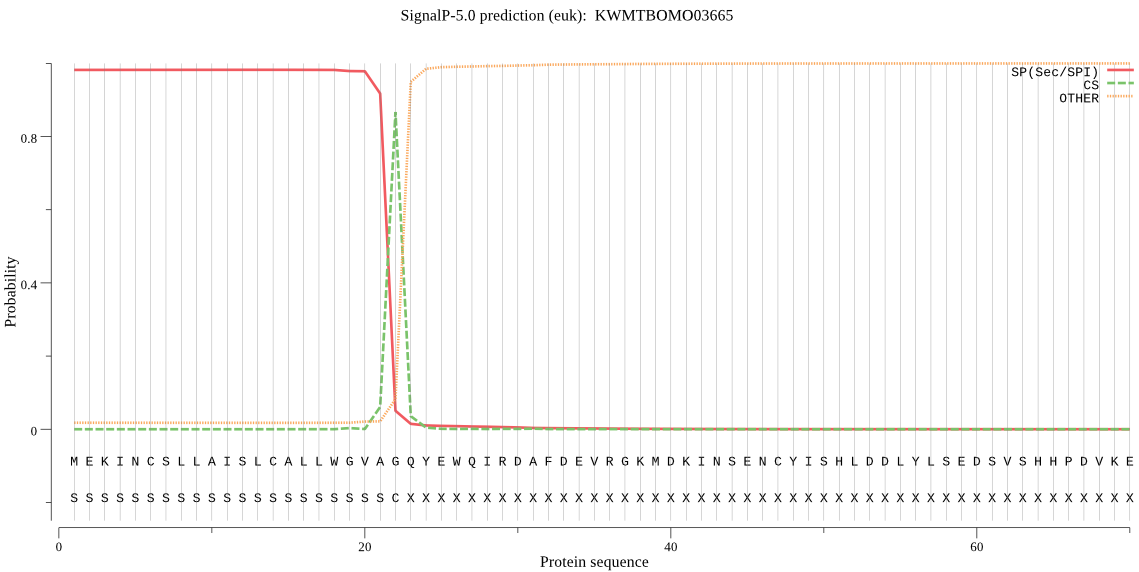
SignalP
Position: 1 - 22,
Likelihood: 0.982260
Length:
800
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
35.57536
Exp number, first 60 AAs:
10.83812
Total prob of N-in:
0.55280
POSSIBLE N-term signal
sequence
outside
1 - 776
TMhelix
777 - 799
inside
800 - 800

Population Genetic Test Statistics
Pi
20.451593
Theta
18.006192
Tajima's D
-1.519989
CLR
1.635626
CSRT
0.0570471476426179
Interpretation
Uncertain
