Gene
KWMTBOMO03664
Annotation
PREDICTED:_zinc_finger_protein_235-like_[Bombyx_mori]
Full name
Zinc finger protein 628
Alternative Name
Zinc finger protein expressed in embryonal cells and certain adult organs
Transcription factor
Location in the cell
Mitochondrial Reliability : 1.275 Nuclear Reliability : 1.913
Sequence
CDS
ATGCGCACGCACACGGGCGAGCGACCCTTCCAGTGTCCGGACTGCCCCAAGGGCTTCGTCAAGCAGTCCTCACTCAAGAGCCACTCGACGGTGCACACGAGCGAGCGATCGTTCGCCTGCGAGCTCTGTCCCAAGACCTTCAAGTGGCGGTCCGGTCTGCGCACCCATCAACAAACCGTACATTTTGGTATAAAACTTCCTGCCAGAAGAAAAAACCAACCTCAAGTCGCAGTTCCAACGATTTTCTGA
Protein
MRTHTGERPFQCPDCPKGFVKQSSLKSHSTVHTSERSFACELCPKTFKWRSGLRTHQQTVHFGIKLPARRKNQPQVAVPTIF
Summary
Description
Transcription activator. Binds DNA on GT-box consensus sequence 5'-TTGGTT-3'.
Transcription activator. Binds DNA on GT-box consensus sequence 5'-TTGGTT-3' (By similarity).
Transcription activator. Binds DNA on GT-box consensus sequence 5'-TTGGTT-3' (By similarity).
Keywords
Alternative splicing
Complete proteome
DNA-binding
Metal-binding
Nucleus
Phosphoprotein
Reference proteome
Repeat
Transcription
Transcription regulation
Zinc
Zinc-finger
Feature
chain Zinc finger protein 628
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
H9IY74
A0A0L7KU31
A0A1S3WDQ9
A0A131Y7M8
A0A3B3VEN3
A0A3P8QSX9
+ More
A0A3B5LEA9 A0A3P9N258 F7AX10 A0A0K8RNH0 F7AX04 M4B041 H2LRX9 A0A2D0RS12 A0A131XVA0 A0A3B3WTN9 A0A146RAN4 A0A1S3GME0 A0A3P9MXE9 A0A2D4KJD3 A0A2D4KJD7 A0A224ZCC2 A0A3B4EKJ2 A0A286XC32 A0A1S3RZ37 L9KPP2 A0A060VXR1 Q4T8D2 A0A1S3PW73 A0A2Y9GFQ5 F1ML21 A0A131Y6Q0 A0A1B6IPL3 A0A2B4S713 A0A2U3ZLM6 A0A3M6TKD3 A0A131XV23 A0A131Y2S4 A0A1S3WDL6 V5I4B9 A0A3B3BP63 A0A3P9I7C9 A0A3Q3G4K6 A0A1S3AI42 A0A3B5JZZ4 A0A2K6M1J6 A0A3B5KAY9 A0A147BF01 A0A384A281 F6V0G1 A0A2U3YYR1 A0A146QIW3 A0A3Q1IJS8 A0A1S3SJW8 A0A2D4LWM7 W5NT75 A0A1U7R272 A0A147BF03 A0A2R9AXE0 L5KCU4 Q8CJ78 A0A3P8P0U0 A0A3Q1HJQ5 G1P722 V5IET8 A0A2I0T6I1 A0A383ZKA9 F7GJE6 A0A2K6QUX0 A0A3Q7RFR5 A0A3B4GSX1 A0A2R8M2R2 A0A2K6DHT8 A0A1W4Y7M8 A0A1W4YHA6 W5LJQ6 A0A2K6QV06 A0A1S3NAB3 A0A2J8IWS7 A0A2J8IWS8 A0A2I3TEI2 A0A087WSX5 H0XIR8 F1SS53 A0A2J8R3G3 A0A1D5QGJ5 A0A0A0MU01 Q5EBL2 A0A2K5QIN9 G1RHV2 A0A3Q7V5A6 A0A146QIU8 G3S3A2 A0A2K5NER9 A0A0D9SE62 A0A2J8R3E3 A0A2Y9IH32 A0A3Q2VGA9
A0A3B5LEA9 A0A3P9N258 F7AX10 A0A0K8RNH0 F7AX04 M4B041 H2LRX9 A0A2D0RS12 A0A131XVA0 A0A3B3WTN9 A0A146RAN4 A0A1S3GME0 A0A3P9MXE9 A0A2D4KJD3 A0A2D4KJD7 A0A224ZCC2 A0A3B4EKJ2 A0A286XC32 A0A1S3RZ37 L9KPP2 A0A060VXR1 Q4T8D2 A0A1S3PW73 A0A2Y9GFQ5 F1ML21 A0A131Y6Q0 A0A1B6IPL3 A0A2B4S713 A0A2U3ZLM6 A0A3M6TKD3 A0A131XV23 A0A131Y2S4 A0A1S3WDL6 V5I4B9 A0A3B3BP63 A0A3P9I7C9 A0A3Q3G4K6 A0A1S3AI42 A0A3B5JZZ4 A0A2K6M1J6 A0A3B5KAY9 A0A147BF01 A0A384A281 F6V0G1 A0A2U3YYR1 A0A146QIW3 A0A3Q1IJS8 A0A1S3SJW8 A0A2D4LWM7 W5NT75 A0A1U7R272 A0A147BF03 A0A2R9AXE0 L5KCU4 Q8CJ78 A0A3P8P0U0 A0A3Q1HJQ5 G1P722 V5IET8 A0A2I0T6I1 A0A383ZKA9 F7GJE6 A0A2K6QUX0 A0A3Q7RFR5 A0A3B4GSX1 A0A2R8M2R2 A0A2K6DHT8 A0A1W4Y7M8 A0A1W4YHA6 W5LJQ6 A0A2K6QV06 A0A1S3NAB3 A0A2J8IWS7 A0A2J8IWS8 A0A2I3TEI2 A0A087WSX5 H0XIR8 F1SS53 A0A2J8R3G3 A0A1D5QGJ5 A0A0A0MU01 Q5EBL2 A0A2K5QIN9 G1RHV2 A0A3Q7V5A6 A0A146QIU8 G3S3A2 A0A2K5NER9 A0A0D9SE62 A0A2J8R3E3 A0A2Y9IH32 A0A3Q2VGA9
Pubmed
EMBL
BABH01038386
BABH01038387
BABH01038388
BABH01038389
JTDY01005793
KOB66620.1
+ More
GEFM01001304 JAP74492.1 AAMC01062641 AAMC01062642 GADI01001345 JAA72463.1 GEFM01006295 JAP69501.1 GCES01120940 JAQ65382.1 IACL01072278 LAB08794.1 IACL01072281 LAB08798.1 GFPF01013534 MAA24680.1 AAKN02046214 KB320717 ELW64756.1 FR904333 CDQ59788.1 CAAE01007830 CAF90850.1 GEFM01001624 JAP74172.1 GECU01018863 JAS88843.1 LSMT01000179 PFX24338.1 RCHS01003437 RMX41806.1 GEFM01006290 JAP69506.1 GEFM01003605 JAP72191.1 GANP01001380 JAB83088.1 GEGO01006037 JAR89367.1 GCES01130545 JAQ55777.1 IACM01045508 LAB25507.1 AMGL01029757 AMGL01029758 AMGL01029759 AMGL01029760 GEGO01006392 JAR89012.1 AJFE02020318 AJFE02020319 KB030840 ELK09167.1 AF435832 AK155214 BC056945 BC062973 AAPE02025289 GANP01009612 JAB74856.1 KZ517182 PKU29409.1 JSUE03022374 JSUE03022375 NBAG03000567 PNI14962.1 PNI14963.1 AACZ04002217 AC008735 AAQR03181554 AEMK02000082 NDHI03003771 PNJ03061.1 AHZZ02014979 BC047332 BC089449 ADFV01100199 GCES01130299 JAQ56023.1 CABD030114970 CABD030114971 AQIB01135263 PNJ03059.1
GEFM01001304 JAP74492.1 AAMC01062641 AAMC01062642 GADI01001345 JAA72463.1 GEFM01006295 JAP69501.1 GCES01120940 JAQ65382.1 IACL01072278 LAB08794.1 IACL01072281 LAB08798.1 GFPF01013534 MAA24680.1 AAKN02046214 KB320717 ELW64756.1 FR904333 CDQ59788.1 CAAE01007830 CAF90850.1 GEFM01001624 JAP74172.1 GECU01018863 JAS88843.1 LSMT01000179 PFX24338.1 RCHS01003437 RMX41806.1 GEFM01006290 JAP69506.1 GEFM01003605 JAP72191.1 GANP01001380 JAB83088.1 GEGO01006037 JAR89367.1 GCES01130545 JAQ55777.1 IACM01045508 LAB25507.1 AMGL01029757 AMGL01029758 AMGL01029759 AMGL01029760 GEGO01006392 JAR89012.1 AJFE02020318 AJFE02020319 KB030840 ELK09167.1 AF435832 AK155214 BC056945 BC062973 AAPE02025289 GANP01009612 JAB74856.1 KZ517182 PKU29409.1 JSUE03022374 JSUE03022375 NBAG03000567 PNI14962.1 PNI14963.1 AACZ04002217 AC008735 AAQR03181554 AEMK02000082 NDHI03003771 PNJ03061.1 AHZZ02014979 BC047332 BC089449 ADFV01100199 GCES01130299 JAQ56023.1 CABD030114970 CABD030114971 AQIB01135263 PNJ03059.1
Proteomes
UP000005204
UP000037510
UP000079721
UP000261500
UP000265100
UP000261380
+ More
UP000242638 UP000008143 UP000002852 UP000001038 UP000221080 UP000261480 UP000081671 UP000261440 UP000005447 UP000087266 UP000011518 UP000193380 UP000248481 UP000009136 UP000225706 UP000245340 UP000275408 UP000261560 UP000265200 UP000261660 UP000005226 UP000233180 UP000261681 UP000002280 UP000245341 UP000265040 UP000002356 UP000189706 UP000240080 UP000010552 UP000000589 UP000257200 UP000001074 UP000006718 UP000233200 UP000286640 UP000261460 UP000008225 UP000233120 UP000192224 UP000018467 UP000002277 UP000005640 UP000005225 UP000008227 UP000028761 UP000233040 UP000001073 UP000286642 UP000001519 UP000233060 UP000029965 UP000248482 UP000264840
UP000242638 UP000008143 UP000002852 UP000001038 UP000221080 UP000261480 UP000081671 UP000261440 UP000005447 UP000087266 UP000011518 UP000193380 UP000248481 UP000009136 UP000225706 UP000245340 UP000275408 UP000261560 UP000265200 UP000261660 UP000005226 UP000233180 UP000261681 UP000002280 UP000245341 UP000265040 UP000002356 UP000189706 UP000240080 UP000010552 UP000000589 UP000257200 UP000001074 UP000006718 UP000233200 UP000286640 UP000261460 UP000008225 UP000233120 UP000192224 UP000018467 UP000002277 UP000005640 UP000005225 UP000008227 UP000028761 UP000233040 UP000001073 UP000286642 UP000001519 UP000233060 UP000029965 UP000248482 UP000264840
Pfam
Interpro
IPR036236
Znf_C2H2_sf
+ More
IPR012934 Znf_AD
IPR013087 Znf_C2H2_type
IPR025398 DUF4371
IPR036051 KRAB_dom_sf
IPR001909 KRAB
IPR041697 Znf-C2H2_11
IPR038441 THAP_Znf_sf
IPR006612 THAP_Znf
IPR036772 SRCR-like_dom_sf
IPR001190 SRCR
IPR017448 SRCR-like_dom
IPR003309 SCAN_dom
IPR038269 SCAN_sf
IPR027764 Zfp18
IPR017114 YY-1-like
IPR012934 Znf_AD
IPR013087 Znf_C2H2_type
IPR025398 DUF4371
IPR036051 KRAB_dom_sf
IPR001909 KRAB
IPR041697 Znf-C2H2_11
IPR038441 THAP_Znf_sf
IPR006612 THAP_Znf
IPR036772 SRCR-like_dom_sf
IPR001190 SRCR
IPR017448 SRCR-like_dom
IPR003309 SCAN_dom
IPR038269 SCAN_sf
IPR027764 Zfp18
IPR017114 YY-1-like
Gene 3D
ProteinModelPortal
H9IY74
A0A0L7KU31
A0A1S3WDQ9
A0A131Y7M8
A0A3B3VEN3
A0A3P8QSX9
+ More
A0A3B5LEA9 A0A3P9N258 F7AX10 A0A0K8RNH0 F7AX04 M4B041 H2LRX9 A0A2D0RS12 A0A131XVA0 A0A3B3WTN9 A0A146RAN4 A0A1S3GME0 A0A3P9MXE9 A0A2D4KJD3 A0A2D4KJD7 A0A224ZCC2 A0A3B4EKJ2 A0A286XC32 A0A1S3RZ37 L9KPP2 A0A060VXR1 Q4T8D2 A0A1S3PW73 A0A2Y9GFQ5 F1ML21 A0A131Y6Q0 A0A1B6IPL3 A0A2B4S713 A0A2U3ZLM6 A0A3M6TKD3 A0A131XV23 A0A131Y2S4 A0A1S3WDL6 V5I4B9 A0A3B3BP63 A0A3P9I7C9 A0A3Q3G4K6 A0A1S3AI42 A0A3B5JZZ4 A0A2K6M1J6 A0A3B5KAY9 A0A147BF01 A0A384A281 F6V0G1 A0A2U3YYR1 A0A146QIW3 A0A3Q1IJS8 A0A1S3SJW8 A0A2D4LWM7 W5NT75 A0A1U7R272 A0A147BF03 A0A2R9AXE0 L5KCU4 Q8CJ78 A0A3P8P0U0 A0A3Q1HJQ5 G1P722 V5IET8 A0A2I0T6I1 A0A383ZKA9 F7GJE6 A0A2K6QUX0 A0A3Q7RFR5 A0A3B4GSX1 A0A2R8M2R2 A0A2K6DHT8 A0A1W4Y7M8 A0A1W4YHA6 W5LJQ6 A0A2K6QV06 A0A1S3NAB3 A0A2J8IWS7 A0A2J8IWS8 A0A2I3TEI2 A0A087WSX5 H0XIR8 F1SS53 A0A2J8R3G3 A0A1D5QGJ5 A0A0A0MU01 Q5EBL2 A0A2K5QIN9 G1RHV2 A0A3Q7V5A6 A0A146QIU8 G3S3A2 A0A2K5NER9 A0A0D9SE62 A0A2J8R3E3 A0A2Y9IH32 A0A3Q2VGA9
A0A3B5LEA9 A0A3P9N258 F7AX10 A0A0K8RNH0 F7AX04 M4B041 H2LRX9 A0A2D0RS12 A0A131XVA0 A0A3B3WTN9 A0A146RAN4 A0A1S3GME0 A0A3P9MXE9 A0A2D4KJD3 A0A2D4KJD7 A0A224ZCC2 A0A3B4EKJ2 A0A286XC32 A0A1S3RZ37 L9KPP2 A0A060VXR1 Q4T8D2 A0A1S3PW73 A0A2Y9GFQ5 F1ML21 A0A131Y6Q0 A0A1B6IPL3 A0A2B4S713 A0A2U3ZLM6 A0A3M6TKD3 A0A131XV23 A0A131Y2S4 A0A1S3WDL6 V5I4B9 A0A3B3BP63 A0A3P9I7C9 A0A3Q3G4K6 A0A1S3AI42 A0A3B5JZZ4 A0A2K6M1J6 A0A3B5KAY9 A0A147BF01 A0A384A281 F6V0G1 A0A2U3YYR1 A0A146QIW3 A0A3Q1IJS8 A0A1S3SJW8 A0A2D4LWM7 W5NT75 A0A1U7R272 A0A147BF03 A0A2R9AXE0 L5KCU4 Q8CJ78 A0A3P8P0U0 A0A3Q1HJQ5 G1P722 V5IET8 A0A2I0T6I1 A0A383ZKA9 F7GJE6 A0A2K6QUX0 A0A3Q7RFR5 A0A3B4GSX1 A0A2R8M2R2 A0A2K6DHT8 A0A1W4Y7M8 A0A1W4YHA6 W5LJQ6 A0A2K6QV06 A0A1S3NAB3 A0A2J8IWS7 A0A2J8IWS8 A0A2I3TEI2 A0A087WSX5 H0XIR8 F1SS53 A0A2J8R3G3 A0A1D5QGJ5 A0A0A0MU01 Q5EBL2 A0A2K5QIN9 G1RHV2 A0A3Q7V5A6 A0A146QIU8 G3S3A2 A0A2K5NER9 A0A0D9SE62 A0A2J8R3E3 A0A2Y9IH32 A0A3Q2VGA9
PDB
3W5K
E-value=3.15894e-09,
Score=140
Ontologies
GO
PANTHER
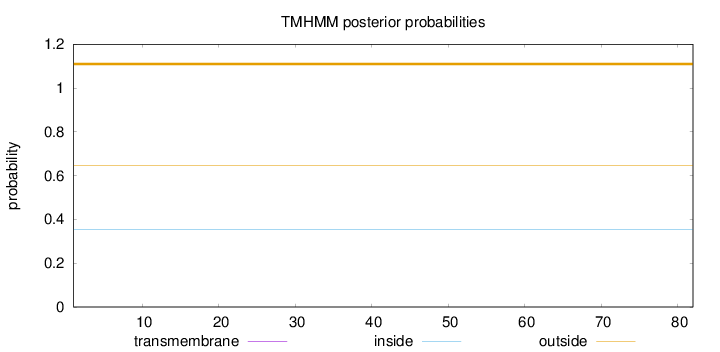
Topology
Subcellular location
Nucleus
Length:
82
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.35509
outside
1 - 82
Population Genetic Test Statistics
Pi
5.361141
Theta
15.773279
Tajima's D
-1.637048
CLR
2.737433
CSRT
0.0433478326083696
Interpretation
Uncertain
