Pre Gene Modal
BGIBMGA002225
Annotation
PREDICTED:_mitochondrial_pyruvate_carrier_1-like_[Plutella_xylostella]
Full name
Mitochondrial pyruvate carrier
Location in the cell
Mitochondrial Reliability : 1.879 PlasmaMembrane Reliability : 1.239
Sequence
CDS
ATGAGTGCTTTAGCAAGGAAACTAGTGAACCAGCTGAAAAGCAAGGAGTTCAGGGACTATTTAATGAGCACACACTTCTGGGGCCCTGTTGCCAATTGGGGTATACCACTAGCAGCCATAGCAGATACAAGGAAGGATCCAAATTTTATAAGTGGAAAAATGACTTTTGCCCTGTCTCTCTACTCGCTGATGTTCATGAGGTTCGCTTGGAAGGTCCAGCCGAGGAACCTGTTGCTCTTCGCCTGTCATTTTACGAACGAATGCGCTCAGCTCACCCAAGGAGCCAGGTTTATAAACTACCACTACATCCAAGGTCCTAAAGAGGTCCAAGAAAAGAAAAGCCAATGA
Protein
MSALARKLVNQLKSKEFRDYLMSTHFWGPVANWGIPLAAIADTRKDPNFISGKMTFALSLYSLMFMRFAWKVQPRNLLLFACHFTNECAQLTQGARFINYHYIQGPKEVQEKKSQ
Summary
Description
Mediates the uptake of pyruvate into mitochondria.
Similarity
Belongs to the mitochondrial pyruvate carrier (MPC) (TC 2.A.105) family.
Uniprot
H9IY92
A0A0L7LM66
A0A194QG24
A0A194R8V3
A0A3S2KY24
A0A1B6KKW9
+ More
A0A069DNC0 A0A2A4IS31 A0A0P4VXI5 A0A2A4IT72 A0A1D2MG28 R4UN71 A0A023F556 A0A1Y1JZS9 D6WCC1 A0A1B6E4A6 A0A067RGP4 A0A182UX89 A0A2R5LGP0 A0A182M343 A0A182L952 A0A182X9V7 Q7PXI3 A0A182HKU0 A0A182K9W6 A0A2M4C4Q2 A0A182FP26 A0A2M4AYV3 A0A2P8ZIL9 A0A2M3ZAX3 A0A182N5F7 A0A182VRA9 A0A182QXD2 A0A182Y578 A0A0K8TP74 A0A182RPL8 V5G9Q9 E0VKM9 A0A0T6AUP8 A0A0L8HC22 T1DSL5 W5JWF6 A0A0A1WLB4 A0A1B0DPV9 A0A182UI38 A0A023GEF8 G3MQS4 A0A2S2NB04 A0A182P2E7 A0A034WAI3 A0A212EI52 C4WY74 A0A1L8DZW1 A0A1Q3FI58 A0A0K8U6A6 A0A182J9V4 A0A0K8RLQ8 B7P1L8 A0A023FT51 A0A0A9XKN3 A0A1B6GZ74 A0A0P4W6U5 A0A182G8A5 A0A2H8TYU5 A0A023FDT1 W8BXJ9 B0W9C4 U5EEU3 A0A0C9RGT2 A0A1Y1YST9 A0A131YW47 A0A336L601 A0A0L0CK09 A0A1W4ZW72 A0A1B6J1M1 Q6DGZ7 A0A1L8EGL0 Q6DD97 A0A0M8ZVW1 Q5M994 A0A1J1I370 F1Q6Z3 F7ATW8 A0A1W4ZM32 E3TFX4 Q17GV8 A0A151JRV0 A0A0C9RS02 A0A2C9K1I1 C1BG19 B5X6D4 A0A0C9RUQ6 A0A2S2QWZ2 A0A0E9X1H1 B5X637 A0A2A3EE82 V9IFJ7 A0A088AN56 B5X6Z2 A0A0C9QYS5
A0A069DNC0 A0A2A4IS31 A0A0P4VXI5 A0A2A4IT72 A0A1D2MG28 R4UN71 A0A023F556 A0A1Y1JZS9 D6WCC1 A0A1B6E4A6 A0A067RGP4 A0A182UX89 A0A2R5LGP0 A0A182M343 A0A182L952 A0A182X9V7 Q7PXI3 A0A182HKU0 A0A182K9W6 A0A2M4C4Q2 A0A182FP26 A0A2M4AYV3 A0A2P8ZIL9 A0A2M3ZAX3 A0A182N5F7 A0A182VRA9 A0A182QXD2 A0A182Y578 A0A0K8TP74 A0A182RPL8 V5G9Q9 E0VKM9 A0A0T6AUP8 A0A0L8HC22 T1DSL5 W5JWF6 A0A0A1WLB4 A0A1B0DPV9 A0A182UI38 A0A023GEF8 G3MQS4 A0A2S2NB04 A0A182P2E7 A0A034WAI3 A0A212EI52 C4WY74 A0A1L8DZW1 A0A1Q3FI58 A0A0K8U6A6 A0A182J9V4 A0A0K8RLQ8 B7P1L8 A0A023FT51 A0A0A9XKN3 A0A1B6GZ74 A0A0P4W6U5 A0A182G8A5 A0A2H8TYU5 A0A023FDT1 W8BXJ9 B0W9C4 U5EEU3 A0A0C9RGT2 A0A1Y1YST9 A0A131YW47 A0A336L601 A0A0L0CK09 A0A1W4ZW72 A0A1B6J1M1 Q6DGZ7 A0A1L8EGL0 Q6DD97 A0A0M8ZVW1 Q5M994 A0A1J1I370 F1Q6Z3 F7ATW8 A0A1W4ZM32 E3TFX4 Q17GV8 A0A151JRV0 A0A0C9RS02 A0A2C9K1I1 C1BG19 B5X6D4 A0A0C9RUQ6 A0A2S2QWZ2 A0A0E9X1H1 B5X637 A0A2A3EE82 V9IFJ7 A0A088AN56 B5X6Z2 A0A0C9QYS5
Pubmed
19121390
26227816
26354079
26334808
27129103
27289101
+ More
25474469 28004739 18362917 19820115 24845553 20966253 12364791 14747013 17210077 29403074 25244985 26369729 20566863 20920257 23761445 25830018 22216098 25348373 22118469 25401762 26823975 26483478 24495485 26830274 26108605 27762356 23594743 20431018 20634964 17510324 15562597 20433749 25613341 26131772
25474469 28004739 18362917 19820115 24845553 20966253 12364791 14747013 17210077 29403074 25244985 26369729 20566863 20920257 23761445 25830018 22216098 25348373 22118469 25401762 26823975 26483478 24495485 26830274 26108605 27762356 23594743 20431018 20634964 17510324 15562597 20433749 25613341 26131772
EMBL
BABH01038392
BABH01038393
JTDY01000596
KOB76520.1
KQ459011
KPJ04364.1
+ More
KQ460500 KPJ14268.1 RSAL01000828 RVE41122.1 GEBQ01027886 JAT12091.1 GBGD01003574 JAC85315.1 NWSH01008362 PCG62585.1 GDKW01002301 JAI54294.1 PCG62588.1 LJIJ01001377 ODM91929.1 KC740771 AGM32595.1 GBBI01002509 JAC16203.1 GEZM01098752 JAV53771.1 KQ971316 EEZ97817.1 GEDC01004522 JAS32776.1 KK852517 KDR22173.1 GGLE01004502 MBY08628.1 AXCM01014955 AAAB01008987 EAA01075.4 APCN01000869 GGFJ01010817 MBW59958.1 GGFK01012636 MBW45957.1 PYGN01000045 PSN56349.1 GGFM01004912 MBW25663.1 AXCN02001173 GDAI01001421 JAI16182.1 GALX01001601 JAB66865.1 DS235250 EEB13935.1 LJIG01022807 KRT78613.1 KQ418693 KOF86315.1 GAMD01001424 JAB00167.1 ADMH02000064 ETN67943.1 GBXI01014493 JAC99798.1 AJVK01018635 AJVK01018636 AJVK01018637 GBBM01003146 JAC32272.1 JO844225 AEO35842.1 GGMR01001688 MBY14307.1 GAKP01008160 GAKP01008159 JAC50793.1 AGBW02014704 OWR41158.1 ABLF02034389 AK343065 BAH72844.1 GFDF01002148 JAV11936.1 GFDL01007820 JAV27225.1 GDHF01030223 GDHF01007435 JAI22091.1 JAI44879.1 GADI01001787 JAA72021.1 ABJB010369566 DS616928 EEC00490.1 GBBL01002369 JAC24951.1 GBHO01022318 GBHO01022317 GBRD01002288 GDHC01020513 GDHC01009386 JAG21286.1 JAG21287.1 JAG63533.1 JAP98115.1 JAQ09243.1 GECZ01002043 JAS67726.1 GDRN01087393 JAI61041.1 JXUM01047423 JXUM01047424 KQ561518 KXJ78310.1 GFXV01006523 MBW18328.1 GBBK01004396 GBBK01004395 JAC20086.1 GAMC01002544 GAMC01002543 JAC04013.1 DS231863 EDS39984.1 GANO01004163 JAB55708.1 GBYB01012362 JAG82129.1 MCFE01000074 ORY01081.1 GEDV01005817 JAP82740.1 UFQS01000111 UFQS01002019 UFQT01000111 UFQT01002019 SSW99824.1 SSX12980.1 SSX20204.1 JRES01000280 KNC32738.1 GECU01026027 GECU01014627 JAS81679.1 JAS93079.1 BC076187 AAH76187.1 GFDG01000952 JAV17847.1 BC077699 CR760678 AAH77699.1 CAJ82200.1 KQ435824 KOX72200.1 BC087474 CM004475 AAH87474.1 OCT77989.1 CVRI01000039 CRK94643.1 CU694274 AAMC01124593 AAMC01124594 GU589259 ADO29210.1 CH477255 EAT45897.1 KQ978566 KYN30087.1 GBYB01011360 JAG81127.1 BT073548 ACO07972.1 BT046603 BT047092 BT048017 ACI66404.1 ACI66893.1 ACI67818.1 GBYB01011361 GBYB01011362 GBYB01011363 JAG81128.1 JAG81129.1 JAG81130.1 GGMS01013052 MBY82255.1 GBXM01012291 JAH96286.1 BT046506 ACI66307.1 KZ288269 PBC30020.1 JR044414 AEY59888.1 BT046811 ACI66612.1 GBZX01002820 JAG89920.1
KQ460500 KPJ14268.1 RSAL01000828 RVE41122.1 GEBQ01027886 JAT12091.1 GBGD01003574 JAC85315.1 NWSH01008362 PCG62585.1 GDKW01002301 JAI54294.1 PCG62588.1 LJIJ01001377 ODM91929.1 KC740771 AGM32595.1 GBBI01002509 JAC16203.1 GEZM01098752 JAV53771.1 KQ971316 EEZ97817.1 GEDC01004522 JAS32776.1 KK852517 KDR22173.1 GGLE01004502 MBY08628.1 AXCM01014955 AAAB01008987 EAA01075.4 APCN01000869 GGFJ01010817 MBW59958.1 GGFK01012636 MBW45957.1 PYGN01000045 PSN56349.1 GGFM01004912 MBW25663.1 AXCN02001173 GDAI01001421 JAI16182.1 GALX01001601 JAB66865.1 DS235250 EEB13935.1 LJIG01022807 KRT78613.1 KQ418693 KOF86315.1 GAMD01001424 JAB00167.1 ADMH02000064 ETN67943.1 GBXI01014493 JAC99798.1 AJVK01018635 AJVK01018636 AJVK01018637 GBBM01003146 JAC32272.1 JO844225 AEO35842.1 GGMR01001688 MBY14307.1 GAKP01008160 GAKP01008159 JAC50793.1 AGBW02014704 OWR41158.1 ABLF02034389 AK343065 BAH72844.1 GFDF01002148 JAV11936.1 GFDL01007820 JAV27225.1 GDHF01030223 GDHF01007435 JAI22091.1 JAI44879.1 GADI01001787 JAA72021.1 ABJB010369566 DS616928 EEC00490.1 GBBL01002369 JAC24951.1 GBHO01022318 GBHO01022317 GBRD01002288 GDHC01020513 GDHC01009386 JAG21286.1 JAG21287.1 JAG63533.1 JAP98115.1 JAQ09243.1 GECZ01002043 JAS67726.1 GDRN01087393 JAI61041.1 JXUM01047423 JXUM01047424 KQ561518 KXJ78310.1 GFXV01006523 MBW18328.1 GBBK01004396 GBBK01004395 JAC20086.1 GAMC01002544 GAMC01002543 JAC04013.1 DS231863 EDS39984.1 GANO01004163 JAB55708.1 GBYB01012362 JAG82129.1 MCFE01000074 ORY01081.1 GEDV01005817 JAP82740.1 UFQS01000111 UFQS01002019 UFQT01000111 UFQT01002019 SSW99824.1 SSX12980.1 SSX20204.1 JRES01000280 KNC32738.1 GECU01026027 GECU01014627 JAS81679.1 JAS93079.1 BC076187 AAH76187.1 GFDG01000952 JAV17847.1 BC077699 CR760678 AAH77699.1 CAJ82200.1 KQ435824 KOX72200.1 BC087474 CM004475 AAH87474.1 OCT77989.1 CVRI01000039 CRK94643.1 CU694274 AAMC01124593 AAMC01124594 GU589259 ADO29210.1 CH477255 EAT45897.1 KQ978566 KYN30087.1 GBYB01011360 JAG81127.1 BT073548 ACO07972.1 BT046603 BT047092 BT048017 ACI66404.1 ACI66893.1 ACI67818.1 GBYB01011361 GBYB01011362 GBYB01011363 JAG81128.1 JAG81129.1 JAG81130.1 GGMS01013052 MBY82255.1 GBXM01012291 JAH96286.1 BT046506 ACI66307.1 KZ288269 PBC30020.1 JR044414 AEY59888.1 BT046811 ACI66612.1 GBZX01002820 JAG89920.1
Proteomes
UP000005204
UP000037510
UP000053268
UP000053240
UP000283053
UP000218220
+ More
UP000094527 UP000007266 UP000027135 UP000075903 UP000075883 UP000075882 UP000076407 UP000007062 UP000075840 UP000075881 UP000069272 UP000245037 UP000075884 UP000075920 UP000075886 UP000076408 UP000075900 UP000009046 UP000053454 UP000000673 UP000092462 UP000075902 UP000075885 UP000007151 UP000007819 UP000075880 UP000001555 UP000069940 UP000249989 UP000002320 UP000193498 UP000037069 UP000192224 UP000053105 UP000186698 UP000183832 UP000000437 UP000008143 UP000221080 UP000008820 UP000078492 UP000076420 UP000087266 UP000242457 UP000005203
UP000094527 UP000007266 UP000027135 UP000075903 UP000075883 UP000075882 UP000076407 UP000007062 UP000075840 UP000075881 UP000069272 UP000245037 UP000075884 UP000075920 UP000075886 UP000076408 UP000075900 UP000009046 UP000053454 UP000000673 UP000092462 UP000075902 UP000075885 UP000007151 UP000007819 UP000075880 UP000001555 UP000069940 UP000249989 UP000002320 UP000193498 UP000037069 UP000192224 UP000053105 UP000186698 UP000183832 UP000000437 UP000008143 UP000221080 UP000008820 UP000078492 UP000076420 UP000087266 UP000242457 UP000005203
PRIDE
Pfam
PF03650 MPC
Interpro
IPR005336
MPC
ProteinModelPortal
H9IY92
A0A0L7LM66
A0A194QG24
A0A194R8V3
A0A3S2KY24
A0A1B6KKW9
+ More
A0A069DNC0 A0A2A4IS31 A0A0P4VXI5 A0A2A4IT72 A0A1D2MG28 R4UN71 A0A023F556 A0A1Y1JZS9 D6WCC1 A0A1B6E4A6 A0A067RGP4 A0A182UX89 A0A2R5LGP0 A0A182M343 A0A182L952 A0A182X9V7 Q7PXI3 A0A182HKU0 A0A182K9W6 A0A2M4C4Q2 A0A182FP26 A0A2M4AYV3 A0A2P8ZIL9 A0A2M3ZAX3 A0A182N5F7 A0A182VRA9 A0A182QXD2 A0A182Y578 A0A0K8TP74 A0A182RPL8 V5G9Q9 E0VKM9 A0A0T6AUP8 A0A0L8HC22 T1DSL5 W5JWF6 A0A0A1WLB4 A0A1B0DPV9 A0A182UI38 A0A023GEF8 G3MQS4 A0A2S2NB04 A0A182P2E7 A0A034WAI3 A0A212EI52 C4WY74 A0A1L8DZW1 A0A1Q3FI58 A0A0K8U6A6 A0A182J9V4 A0A0K8RLQ8 B7P1L8 A0A023FT51 A0A0A9XKN3 A0A1B6GZ74 A0A0P4W6U5 A0A182G8A5 A0A2H8TYU5 A0A023FDT1 W8BXJ9 B0W9C4 U5EEU3 A0A0C9RGT2 A0A1Y1YST9 A0A131YW47 A0A336L601 A0A0L0CK09 A0A1W4ZW72 A0A1B6J1M1 Q6DGZ7 A0A1L8EGL0 Q6DD97 A0A0M8ZVW1 Q5M994 A0A1J1I370 F1Q6Z3 F7ATW8 A0A1W4ZM32 E3TFX4 Q17GV8 A0A151JRV0 A0A0C9RS02 A0A2C9K1I1 C1BG19 B5X6D4 A0A0C9RUQ6 A0A2S2QWZ2 A0A0E9X1H1 B5X637 A0A2A3EE82 V9IFJ7 A0A088AN56 B5X6Z2 A0A0C9QYS5
A0A069DNC0 A0A2A4IS31 A0A0P4VXI5 A0A2A4IT72 A0A1D2MG28 R4UN71 A0A023F556 A0A1Y1JZS9 D6WCC1 A0A1B6E4A6 A0A067RGP4 A0A182UX89 A0A2R5LGP0 A0A182M343 A0A182L952 A0A182X9V7 Q7PXI3 A0A182HKU0 A0A182K9W6 A0A2M4C4Q2 A0A182FP26 A0A2M4AYV3 A0A2P8ZIL9 A0A2M3ZAX3 A0A182N5F7 A0A182VRA9 A0A182QXD2 A0A182Y578 A0A0K8TP74 A0A182RPL8 V5G9Q9 E0VKM9 A0A0T6AUP8 A0A0L8HC22 T1DSL5 W5JWF6 A0A0A1WLB4 A0A1B0DPV9 A0A182UI38 A0A023GEF8 G3MQS4 A0A2S2NB04 A0A182P2E7 A0A034WAI3 A0A212EI52 C4WY74 A0A1L8DZW1 A0A1Q3FI58 A0A0K8U6A6 A0A182J9V4 A0A0K8RLQ8 B7P1L8 A0A023FT51 A0A0A9XKN3 A0A1B6GZ74 A0A0P4W6U5 A0A182G8A5 A0A2H8TYU5 A0A023FDT1 W8BXJ9 B0W9C4 U5EEU3 A0A0C9RGT2 A0A1Y1YST9 A0A131YW47 A0A336L601 A0A0L0CK09 A0A1W4ZW72 A0A1B6J1M1 Q6DGZ7 A0A1L8EGL0 Q6DD97 A0A0M8ZVW1 Q5M994 A0A1J1I370 F1Q6Z3 F7ATW8 A0A1W4ZM32 E3TFX4 Q17GV8 A0A151JRV0 A0A0C9RS02 A0A2C9K1I1 C1BG19 B5X6D4 A0A0C9RUQ6 A0A2S2QWZ2 A0A0E9X1H1 B5X637 A0A2A3EE82 V9IFJ7 A0A088AN56 B5X6Z2 A0A0C9QYS5
Ontologies
GO
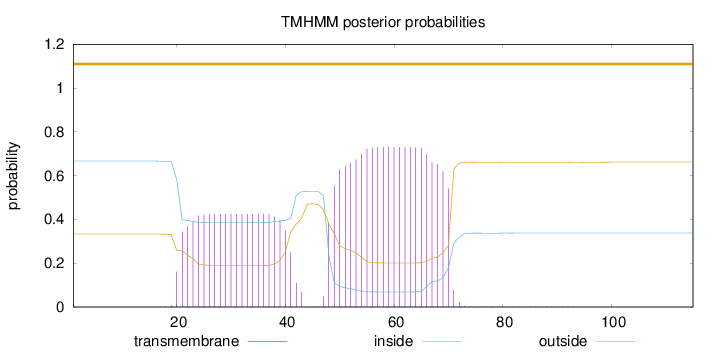
Topology
Subcellular location
Mitochondrion inner membrane
Length:
115
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
24.44664
Exp number, first 60 AAs:
17.47739
Total prob of N-in:
0.66680
POSSIBLE N-term signal
sequence
outside
1 - 115
Population Genetic Test Statistics
Pi
1.857356
Theta
7.831134
Tajima's D
-2.328349
CLR
71.509498
CSRT
0.00194990250487476
Interpretation
Uncertain
