Gene
KWMTBOMO03656 Validated by peptides from experiments
Annotation
PREDICTED:_ras-related_protein_Rab-18-like_[Papilio_xuthus]
Full name
Ras-related protein Rab-18
Location in the cell
Cytoplasmic Reliability : 3.015
Sequence
CDS
ATGGATTGTGTGGCTAAATTGAAAATTCTGTTGCTTGGTGAGAGTGGCGTTGGTAAATCTAGTATTATATTAGCGTTCACAAAAGGCGACTACCAAGCACAGTTCCCTGCTACAATCGGAGTGGACTACAACCATAAGATTATGGATGTAAATGGACTAAAAGTTAAACTCGGAATTTGGGATACGGCAGGACAAGAACGATATAGAACTTTAACAACCAGTTTTTACAGAGAAGCGCACGGTGCTATATTAGTTTATGACGTATCTGAACCAAAAACCTTAGCAAAGTTAAACGAATGGGTTGAAGAGTTACAAGTTTACACTACGAATAAGAATATTGTCTGTTTGGTCGTTGGAAATAAAATTGATAAGGATAGAGCGGTACCCCGTGAACACGGTCAGGCCTTCGCTCAGAAACACAGAATGCTTTTCATCGAGAGCAGTGCTAAAACACAAGAAGGCATACATCTCGCCTTTGAGGAACTGACCCAAAAGATTATCGAAACTCCTGGATTATGGGAGAAAGACACATCGGCTTCAAACATTCGGGTTACAGAGGAAGAAGCGAGAGAACAAGGTGGATCGTGCTACGAAACAACTTGTTCCATAAAATAA
Protein
MDCVAKLKILLLGESGVGKSSIILAFTKGDYQAQFPATIGVDYNHKIMDVNGLKVKLGIWDTAGQERYRTLTTSFYREAHGAILVYDVSEPKTLAKLNEWVEELQVYTTNKNIVCLVVGNKIDKDRAVPREHGQAFAQKHRMLFIESSAKTQEGIHLAFEELTQKIIETPGLWEKDTSASNIRVTEEEAREQGGSCYETTCSIK
Summary
Description
Plays a role in apical endocytosis/recycling. May be implicated in transport between the plasma membrane and early endosomes. Plays a key role in eye and brain development and neurodegeneration (By similarity).
Similarity
Belongs to the small GTPase superfamily. Rab family.
Keywords
Acetylation
Cell membrane
Complete proteome
Developmental protein
GTP-binding
Lipoprotein
Membrane
Methylation
Nucleotide-binding
Palmitate
Phosphoprotein
Prenylation
Protein transport
Reference proteome
Transport
Feature
chain Ras-related protein Rab-18
propeptide Removed in mature form
propeptide Removed in mature form
Uniprot
A0A194QFU9
S4NSQ9
A0A2H1W832
A0A2A4JCG2
H9IY70
A0A0P5TEN8
+ More
A0A0P6I0E2 Q4SP01 A0A182U381 H3CR18 A0A182VI95 A0A182X412 Q7QGS6 A0A182I5I1 A0A2M3Z771 A0A1D2MLI6 A0A2M4AEF2 A0A2M4BZY9 W5JHW0 A0A182FKC3 E9HIT2 Q6P129 A0A2M4ADU3 T1L4D2 A0A067R6Y7 Q6UYC2 A0A182JCY5 A0A224XKC4 A0A069DQ33 Q0PD38 P35293 A0A3Q0DBU1 G3H3P1 A0A0V0G3Q6 A0A3Q3MIQ1 A0A182PJB0 A0A2Y9E3D5 A0A250YCA0 A0A2K5KNJ8 Q5EB77 A0A1B6I787 A0A1B6FI41 A0A1S3F0L0 I3MN66 A0A286XYW4 A0A0A9XZW7 S4RKZ6 G1TFB5 W5ML35 A0A084VSJ3 G3PVK7 A0A3Q0RKF1 Q28D30 A0A1S2ZX50 A0A182YKM2 A0A182KCB3 A0A1S3Q7V2 A0A182NC92 F6ZX76 A0A1B6KBP7 V9LD26 G3W037 A0A182MC58 A0A061I5P2 A0A3Q3IQ05 A0A060X2Z0 A0A1Q3FHQ6 A0A3B4EXZ0 A0A3Q3CI90 A0A3P8Q0A4 A0A3P9BM22 A0A182QMF7 A0A3Q3FZ10 A0A3Q4MHH7 I3KQW0 A0A3B5BJQ7 A0A0P6IVG1 A0A1D5R5M1 A0A3P8U173 A0A3Q1BJA5 A0A182HB16 H3AT07 A0A226EPC4 Q6GM52 A0A3B3SCQ0 H2SHQ6 T1HYX3 A0A3Q3SBI4 G1KE45 A0A2I4CAC4 A0A3Q1GN16 B0WBF2 E6ZI64 A0A3B3WV80 A0A087Y666 A0A3B3UQZ0 A0A182H3F1 Q7ZY33 A0A315V1Y0
A0A0P6I0E2 Q4SP01 A0A182U381 H3CR18 A0A182VI95 A0A182X412 Q7QGS6 A0A182I5I1 A0A2M3Z771 A0A1D2MLI6 A0A2M4AEF2 A0A2M4BZY9 W5JHW0 A0A182FKC3 E9HIT2 Q6P129 A0A2M4ADU3 T1L4D2 A0A067R6Y7 Q6UYC2 A0A182JCY5 A0A224XKC4 A0A069DQ33 Q0PD38 P35293 A0A3Q0DBU1 G3H3P1 A0A0V0G3Q6 A0A3Q3MIQ1 A0A182PJB0 A0A2Y9E3D5 A0A250YCA0 A0A2K5KNJ8 Q5EB77 A0A1B6I787 A0A1B6FI41 A0A1S3F0L0 I3MN66 A0A286XYW4 A0A0A9XZW7 S4RKZ6 G1TFB5 W5ML35 A0A084VSJ3 G3PVK7 A0A3Q0RKF1 Q28D30 A0A1S2ZX50 A0A182YKM2 A0A182KCB3 A0A1S3Q7V2 A0A182NC92 F6ZX76 A0A1B6KBP7 V9LD26 G3W037 A0A182MC58 A0A061I5P2 A0A3Q3IQ05 A0A060X2Z0 A0A1Q3FHQ6 A0A3B4EXZ0 A0A3Q3CI90 A0A3P8Q0A4 A0A3P9BM22 A0A182QMF7 A0A3Q3FZ10 A0A3Q4MHH7 I3KQW0 A0A3B5BJQ7 A0A0P6IVG1 A0A1D5R5M1 A0A3P8U173 A0A3Q1BJA5 A0A182HB16 H3AT07 A0A226EPC4 Q6GM52 A0A3B3SCQ0 H2SHQ6 T1HYX3 A0A3Q3SBI4 G1KE45 A0A2I4CAC4 A0A3Q1GN16 B0WBF2 E6ZI64 A0A3B3WV80 A0A087Y666 A0A3B3UQZ0 A0A182H3F1 Q7ZY33 A0A315V1Y0
Pubmed
26354079
23622113
19121390
15496914
12364791
14747013
+ More
17210077 27289101 20920257 23761445 21292972 23594743 24845553 26334808 11773082 11856727 12040188 12578829 16923123 21183079 7706395 7916717 16141072 15489334 1555775 26319212 21804562 29704459 28087693 21993624 25401762 26823975 24438588 20431018 25244985 17495919 24402279 21709235 23929341 24755649 25186727 26999592 17431167 26483478 9215903 27762356 29240929 21551351 29703783
17210077 27289101 20920257 23761445 21292972 23594743 24845553 26334808 11773082 11856727 12040188 12578829 16923123 21183079 7706395 7916717 16141072 15489334 1555775 26319212 21804562 29704459 28087693 21993624 25401762 26823975 24438588 20431018 25244985 17495919 24402279 21709235 23929341 24755649 25186727 26999592 17431167 26483478 9215903 27762356 29240929 21551351 29703783
EMBL
KQ459011
KPJ04362.1
GAIX01013967
JAA78593.1
ODYU01006877
SOQ49116.1
+ More
NWSH01002073 PCG69244.1 BABH01038450 GDIP01128964 LRGB01002901 JAL74750.1 KZS05563.1 GDIQ01011390 JAN83347.1 CAAE01014542 CAF97631.1 AAAB01008823 EAA05523.4 APCN01003445 GGFM01003600 MBW24351.1 LJIJ01000902 ODM93857.1 GGFK01005667 MBW38988.1 GGFJ01009494 MBW58635.1 ADMH02001144 ETN63947.1 GL732657 EFX68330.1 BX465854 BC065318 BC153660 AAH65318.1 AAI53661.1 GGFK01005635 MBW38956.1 CAEY01001080 KK852902 KDR14057.1 AY357728 AAQ56773.1 GFTR01003499 JAW12927.1 GBGD01002939 JAC85950.1 AB232612 CH466592 BAF02874.1 EDL23028.1 X80333 L04966 AK045514 AK140901 AK143673 AK146177 AK146397 AK154920 BC056351 M79308 JH000129 RAZU01000120 EGW05179.1 RLQ71808.1 GECL01003439 JAP02685.1 GFFW01003598 JAV41190.1 BC089957 GECU01031637 GECU01029152 GECU01024939 GECU01006274 GECU01001746 JAS76069.1 JAS78554.1 JAS82767.1 JAT01433.1 JAT05961.1 GECZ01019898 JAS49871.1 AGTP01076469 AGTP01076470 AAKN02037547 GBHO01018150 GBRD01006772 GDHC01009809 JAG25454.1 JAG59049.1 JAQ08820.1 AAGW02059558 AAGW02059559 AAGW02059560 AHAT01027019 ATLV01016042 KE525054 KFB40937.1 AAMC01019722 BC135986 CR855736 AAI35987.1 CAJ81541.1 GEBQ01031115 JAT08862.1 JW877730 AFP10247.1 AEFK01200006 AEFK01200007 AEFK01200008 AXCM01005795 KE673613 ERE77110.1 FR904783 CDQ71684.1 GFDL01008002 JAV27043.1 AXCN02001762 AERX01030759 GDUN01000772 JAN95147.1 JSUE03043110 JXUM01123892 KQ566767 KXJ69840.1 AFYH01115001 AFYH01115002 AFYH01115003 AFYH01115004 AFYH01115005 LNIX01000002 OXA59369.1 BC074233 CM004477 AAH74233.1 OCT73930.1 ACPB03021331 DS231879 EDS42391.1 FQ310508 CBN81748.1 AYCK01006041 JXUM01107366 KQ565135 KXJ71298.1 BC043996 AAH43996.1 NHOQ01002364 PWA17503.1
NWSH01002073 PCG69244.1 BABH01038450 GDIP01128964 LRGB01002901 JAL74750.1 KZS05563.1 GDIQ01011390 JAN83347.1 CAAE01014542 CAF97631.1 AAAB01008823 EAA05523.4 APCN01003445 GGFM01003600 MBW24351.1 LJIJ01000902 ODM93857.1 GGFK01005667 MBW38988.1 GGFJ01009494 MBW58635.1 ADMH02001144 ETN63947.1 GL732657 EFX68330.1 BX465854 BC065318 BC153660 AAH65318.1 AAI53661.1 GGFK01005635 MBW38956.1 CAEY01001080 KK852902 KDR14057.1 AY357728 AAQ56773.1 GFTR01003499 JAW12927.1 GBGD01002939 JAC85950.1 AB232612 CH466592 BAF02874.1 EDL23028.1 X80333 L04966 AK045514 AK140901 AK143673 AK146177 AK146397 AK154920 BC056351 M79308 JH000129 RAZU01000120 EGW05179.1 RLQ71808.1 GECL01003439 JAP02685.1 GFFW01003598 JAV41190.1 BC089957 GECU01031637 GECU01029152 GECU01024939 GECU01006274 GECU01001746 JAS76069.1 JAS78554.1 JAS82767.1 JAT01433.1 JAT05961.1 GECZ01019898 JAS49871.1 AGTP01076469 AGTP01076470 AAKN02037547 GBHO01018150 GBRD01006772 GDHC01009809 JAG25454.1 JAG59049.1 JAQ08820.1 AAGW02059558 AAGW02059559 AAGW02059560 AHAT01027019 ATLV01016042 KE525054 KFB40937.1 AAMC01019722 BC135986 CR855736 AAI35987.1 CAJ81541.1 GEBQ01031115 JAT08862.1 JW877730 AFP10247.1 AEFK01200006 AEFK01200007 AEFK01200008 AXCM01005795 KE673613 ERE77110.1 FR904783 CDQ71684.1 GFDL01008002 JAV27043.1 AXCN02001762 AERX01030759 GDUN01000772 JAN95147.1 JSUE03043110 JXUM01123892 KQ566767 KXJ69840.1 AFYH01115001 AFYH01115002 AFYH01115003 AFYH01115004 AFYH01115005 LNIX01000002 OXA59369.1 BC074233 CM004477 AAH74233.1 OCT73930.1 ACPB03021331 DS231879 EDS42391.1 FQ310508 CBN81748.1 AYCK01006041 JXUM01107366 KQ565135 KXJ71298.1 BC043996 AAH43996.1 NHOQ01002364 PWA17503.1
Proteomes
UP000053268
UP000218220
UP000005204
UP000076858
UP000075902
UP000007303
+ More
UP000075903 UP000076407 UP000007062 UP000075840 UP000094527 UP000000673 UP000069272 UP000000305 UP000000437 UP000015104 UP000027135 UP000075880 UP000000589 UP000189706 UP000001075 UP000273346 UP000261640 UP000075885 UP000248480 UP000233060 UP000002494 UP000081671 UP000005215 UP000005447 UP000245300 UP000001811 UP000018468 UP000030765 UP000007635 UP000261340 UP000008143 UP000079721 UP000076408 UP000075881 UP000087266 UP000075884 UP000002280 UP000007648 UP000075883 UP000030759 UP000261600 UP000193380 UP000261460 UP000264840 UP000265100 UP000265160 UP000075886 UP000261660 UP000261580 UP000005207 UP000261400 UP000006718 UP000265080 UP000257160 UP000069940 UP000249989 UP000008672 UP000198287 UP000186698 UP000261540 UP000005226 UP000015103 UP000001646 UP000192220 UP000257200 UP000002320 UP000261480 UP000028760 UP000261500
UP000075903 UP000076407 UP000007062 UP000075840 UP000094527 UP000000673 UP000069272 UP000000305 UP000000437 UP000015104 UP000027135 UP000075880 UP000000589 UP000189706 UP000001075 UP000273346 UP000261640 UP000075885 UP000248480 UP000233060 UP000002494 UP000081671 UP000005215 UP000005447 UP000245300 UP000001811 UP000018468 UP000030765 UP000007635 UP000261340 UP000008143 UP000079721 UP000076408 UP000075881 UP000087266 UP000075884 UP000002280 UP000007648 UP000075883 UP000030759 UP000261600 UP000193380 UP000261460 UP000264840 UP000265100 UP000265160 UP000075886 UP000261660 UP000261580 UP000005207 UP000261400 UP000006718 UP000265080 UP000257160 UP000069940 UP000249989 UP000008672 UP000198287 UP000186698 UP000261540 UP000005226 UP000015103 UP000001646 UP000192220 UP000257200 UP000002320 UP000261480 UP000028760 UP000261500
PRIDE
Pfam
PF00071 Ras
Interpro
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
A0A194QFU9
S4NSQ9
A0A2H1W832
A0A2A4JCG2
H9IY70
A0A0P5TEN8
+ More
A0A0P6I0E2 Q4SP01 A0A182U381 H3CR18 A0A182VI95 A0A182X412 Q7QGS6 A0A182I5I1 A0A2M3Z771 A0A1D2MLI6 A0A2M4AEF2 A0A2M4BZY9 W5JHW0 A0A182FKC3 E9HIT2 Q6P129 A0A2M4ADU3 T1L4D2 A0A067R6Y7 Q6UYC2 A0A182JCY5 A0A224XKC4 A0A069DQ33 Q0PD38 P35293 A0A3Q0DBU1 G3H3P1 A0A0V0G3Q6 A0A3Q3MIQ1 A0A182PJB0 A0A2Y9E3D5 A0A250YCA0 A0A2K5KNJ8 Q5EB77 A0A1B6I787 A0A1B6FI41 A0A1S3F0L0 I3MN66 A0A286XYW4 A0A0A9XZW7 S4RKZ6 G1TFB5 W5ML35 A0A084VSJ3 G3PVK7 A0A3Q0RKF1 Q28D30 A0A1S2ZX50 A0A182YKM2 A0A182KCB3 A0A1S3Q7V2 A0A182NC92 F6ZX76 A0A1B6KBP7 V9LD26 G3W037 A0A182MC58 A0A061I5P2 A0A3Q3IQ05 A0A060X2Z0 A0A1Q3FHQ6 A0A3B4EXZ0 A0A3Q3CI90 A0A3P8Q0A4 A0A3P9BM22 A0A182QMF7 A0A3Q3FZ10 A0A3Q4MHH7 I3KQW0 A0A3B5BJQ7 A0A0P6IVG1 A0A1D5R5M1 A0A3P8U173 A0A3Q1BJA5 A0A182HB16 H3AT07 A0A226EPC4 Q6GM52 A0A3B3SCQ0 H2SHQ6 T1HYX3 A0A3Q3SBI4 G1KE45 A0A2I4CAC4 A0A3Q1GN16 B0WBF2 E6ZI64 A0A3B3WV80 A0A087Y666 A0A3B3UQZ0 A0A182H3F1 Q7ZY33 A0A315V1Y0
A0A0P6I0E2 Q4SP01 A0A182U381 H3CR18 A0A182VI95 A0A182X412 Q7QGS6 A0A182I5I1 A0A2M3Z771 A0A1D2MLI6 A0A2M4AEF2 A0A2M4BZY9 W5JHW0 A0A182FKC3 E9HIT2 Q6P129 A0A2M4ADU3 T1L4D2 A0A067R6Y7 Q6UYC2 A0A182JCY5 A0A224XKC4 A0A069DQ33 Q0PD38 P35293 A0A3Q0DBU1 G3H3P1 A0A0V0G3Q6 A0A3Q3MIQ1 A0A182PJB0 A0A2Y9E3D5 A0A250YCA0 A0A2K5KNJ8 Q5EB77 A0A1B6I787 A0A1B6FI41 A0A1S3F0L0 I3MN66 A0A286XYW4 A0A0A9XZW7 S4RKZ6 G1TFB5 W5ML35 A0A084VSJ3 G3PVK7 A0A3Q0RKF1 Q28D30 A0A1S2ZX50 A0A182YKM2 A0A182KCB3 A0A1S3Q7V2 A0A182NC92 F6ZX76 A0A1B6KBP7 V9LD26 G3W037 A0A182MC58 A0A061I5P2 A0A3Q3IQ05 A0A060X2Z0 A0A1Q3FHQ6 A0A3B4EXZ0 A0A3Q3CI90 A0A3P8Q0A4 A0A3P9BM22 A0A182QMF7 A0A3Q3FZ10 A0A3Q4MHH7 I3KQW0 A0A3B5BJQ7 A0A0P6IVG1 A0A1D5R5M1 A0A3P8U173 A0A3Q1BJA5 A0A182HB16 H3AT07 A0A226EPC4 Q6GM52 A0A3B3SCQ0 H2SHQ6 T1HYX3 A0A3Q3SBI4 G1KE45 A0A2I4CAC4 A0A3Q1GN16 B0WBF2 E6ZI64 A0A3B3WV80 A0A087Y666 A0A3B3UQZ0 A0A182H3F1 Q7ZY33 A0A315V1Y0
PDB
1X3S
E-value=9.49917e-54,
Score=528
Ontologies
GO
Topology
Subcellular location
Apical cell membrane
Basal cell membrane
Basal cell membrane
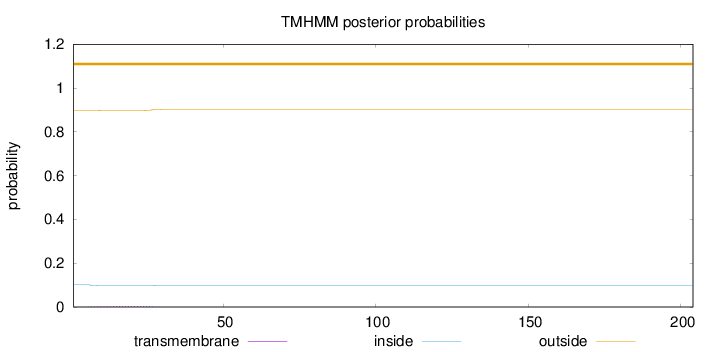
Length:
204
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0907500000000001
Exp number, first 60 AAs:
0.0907500000000001
Total prob of N-in:
0.10156
outside
1 - 204
Population Genetic Test Statistics
Pi
27.049317
Theta
25.491437
Tajima's D
0.213207
CLR
0.915186
CSRT
0.436678166091695
Interpretation
Uncertain
