Gene
KWMTBOMO03648
Pre Gene Modal
BGIBMGA014550
Annotation
desaturase_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 2.781
Sequence
CDS
ATGAAGTTGCTCTTGATGGCTTGCTTCTGCAGCAGTGGCCAGAATTCAATATACAACTGGGTGAGGGATCACAGACTCCATCACAAGTTTGCTGACACCGATGCCGACCCTCACAACGTGAAACGAGGCTTCTTCTTCTCCCACATAGGATGGCTGATGGTGAAGAAGAACGAAAGTGTCCTTACGAAGGGAAAGCTGATTGACATGAGTGACATAGAAAGCGATTCTCATTTGATGTTTGAACATAAATACCACAATCAATTAACAATCCTATTCTGTTTTCTAATACCAACATTAATGAACATCTTCTTGATCGGGGAAGATTGGAAGTGCGCTGTTGCCTGGCAATGCTTCATACGGTATCTCTACGTGCTGCATTGTGAACTGACTGTTAACAGTTTGGCTCATATGTACGGCTATAAACCTTATAACGTAAACATTGAAGCATCAGAGAATATTCTAGTTTCGGTCCTGACGTACGGCGAGGGCTATCACAACTTTCATCATGTCTTCCCATTCGACTTTCGTGCAGCAGAAACTATGGATTTCTTCAGTCTATCCACAAAGATAATCAGTACATTCGAGAAGATTGGTTGGACGTATGACCTCAAGCAAGCCAGTCCAGAAATGATCGAGGCGGCTCGTGACAAGCTAGGGGGTGATGGGTCTAAAGCTCACAATTAA
Protein
MKLLLMACFCSSGQNSIYNWVRDHRLHHKFADTDADPHNVKRGFFFSHIGWLMVKKNESVLTKGKLIDMSDIESDSHLMFEHKYHNQLTILFCFLIPTLMNIFLIGEDWKCAVAWQCFIRYLYVLHCELTVNSLAHMYGYKPYNVNIEASENILVSVLTYGEGYHNFHHVFPFDFRAAETMDFFSLSTKIISTFEKIGWTYDLKQASPEMIEAARDKLGGDGSKAHN
Summary
Cofactor
Fe cation
Similarity
Belongs to the fatty acid desaturase type 1 family.
Uniprot
H9JYD0
G9BY58
A0A2A4JV26
T1RU52
G9BY59
A0A3S2NSR1
+ More
A0A2A4JW88 A0A194REN0 A0A194RIH1 A0A194Q2W5 A0A2H1VHN0 T1RU43 A0A291P0E4 A0A2A4K356 A0A1L8D6H7 T1RTD4 A0A212F8H4 A0A2H1X0N9 Q8MZX4 E3TMU9 A0A1B6DCJ1 B3LZD2 A0A1A9XNV7 A0A1A9VPX3 A0A0J7K2H2 A0A1W4VQC5 B4M6D6 B4NGT2 A0A1B0CEY1 A0A1A9WN17 A0A1W4X6D0 Q8MZY2 A0A1A9ZF92 A0A0P6IHM7 A0A1B0B1J8 A0A1B0GE34 D3TN52 B4R1J8 B4PLU0 Q9VA94 A0A1B6EKR5 A0A232EI48 A0A1I8N2Y4 Q8N001 A0A1Q3F6G7 Q29C24 B4GNP6 A0A1I8PAY4 A0A3B0KVP4 A0A1B6IH88 T1PM50 B4K9T3 W8B9K3 A0A067RJC1 A0A2J7Q0J6 K7J1Y0 A0A2J7Q0J2 K7IYW5 A0A1L8D6I7 A0A0A1X4V7 A0A1W4WVL1 A0A0L0BZP6 A0A067QIN8 A0A182SRI4 A0A034VQK3 Q17CJ9 A0A0K8U130 A0A1Y1LF74 A0A182H3A8 A0A182JFM4 A0A182KA51 A0A1B6MHV3 A0A1S4F5C8 A0A0A9ZCN0 Q17EY7 A0A1S4F557 B0X4F3 A0A3Q0IQ53 B4JEM7 D2A5U2 A0A212EQ61 Q8MZW0 A0A084W6J5 A0A146KPE5 A0A194R858 F5HK94 B3P7X0 A0A182HUB6 A0A182M1I7 B0X4F2 A0A151X2U1 A0A1B6MMW0 W5J5Z7 F4WBY7 A0A151JS66 E2C646 A0A2H4WB77 A0A3S2NP26 J9KA67
A0A2A4JW88 A0A194REN0 A0A194RIH1 A0A194Q2W5 A0A2H1VHN0 T1RU43 A0A291P0E4 A0A2A4K356 A0A1L8D6H7 T1RTD4 A0A212F8H4 A0A2H1X0N9 Q8MZX4 E3TMU9 A0A1B6DCJ1 B3LZD2 A0A1A9XNV7 A0A1A9VPX3 A0A0J7K2H2 A0A1W4VQC5 B4M6D6 B4NGT2 A0A1B0CEY1 A0A1A9WN17 A0A1W4X6D0 Q8MZY2 A0A1A9ZF92 A0A0P6IHM7 A0A1B0B1J8 A0A1B0GE34 D3TN52 B4R1J8 B4PLU0 Q9VA94 A0A1B6EKR5 A0A232EI48 A0A1I8N2Y4 Q8N001 A0A1Q3F6G7 Q29C24 B4GNP6 A0A1I8PAY4 A0A3B0KVP4 A0A1B6IH88 T1PM50 B4K9T3 W8B9K3 A0A067RJC1 A0A2J7Q0J6 K7J1Y0 A0A2J7Q0J2 K7IYW5 A0A1L8D6I7 A0A0A1X4V7 A0A1W4WVL1 A0A0L0BZP6 A0A067QIN8 A0A182SRI4 A0A034VQK3 Q17CJ9 A0A0K8U130 A0A1Y1LF74 A0A182H3A8 A0A182JFM4 A0A182KA51 A0A1B6MHV3 A0A1S4F5C8 A0A0A9ZCN0 Q17EY7 A0A1S4F557 B0X4F3 A0A3Q0IQ53 B4JEM7 D2A5U2 A0A212EQ61 Q8MZW0 A0A084W6J5 A0A146KPE5 A0A194R858 F5HK94 B3P7X0 A0A182HUB6 A0A182M1I7 B0X4F2 A0A151X2U1 A0A1B6MMW0 W5J5Z7 F4WBY7 A0A151JS66 E2C646 A0A2H4WB77 A0A3S2NP26 J9KA67
Pubmed
19121390
24500170
26354079
28986331
22118469
12524345
+ More
20691782 17994087 20353571 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 28648823 25315136 15632085 24495485 24845553 20075255 25830018 26108605 25348373 17510324 28004739 26483478 25401762 18362917 19820115 24438588 26823975 12364791 14747013 17210077 20920257 23761445 21719571 20798317
20691782 17994087 20353571 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 28648823 25315136 15632085 24495485 24845553 20075255 25830018 26108605 25348373 17510324 28004739 26483478 25401762 18362917 19820115 24438588 26823975 12364791 14747013 17210077 20920257 23761445 21719571 20798317
EMBL
BABH01043271
BABH01043272
HQ704865
AET36545.1
NWSH01000559
PCG75659.1
+ More
JX531674 AGO45859.1 HQ704866 AET36546.1 RSAL01000244 RVE43603.1 PCG75660.1 KQ460325 KPJ15745.1 KPJ15746.1 KQ459564 KPI99678.1 ODYU01002605 SOQ40327.1 JX531659 AGO45844.1 MF687531 ATJ44457.1 NWSH01000211 PCG78358.1 GEYN01000134 JAV01995.1 JX531665 AGO45850.1 AGBW02009734 OWR50018.1 ODYU01012535 SOQ58905.1 AF482926 AAM28501.1 GU952764 ADO85597.1 GEDC01030129 GEDC01013889 JAS07169.1 JAS23409.1 CH902617 EDV44111.1 LBMM01016179 KMQ84512.1 CH940652 EDW59212.1 CH964272 EDW84429.1 AJWK01009460 AJWK01009461 AF482918 AAM28493.1 GDIQ01004277 JAN90460.1 JXJN01007190 CCAG010012086 EZ422854 ADD19130.1 CM000364 EDX14986.1 CM000160 EDW99077.1 AE014297 AY047565 AAF57020.1 AAK77297.1 GECZ01031245 JAS38524.1 NNAY01004348 OXU18023.1 AF482899 AAM33369.1 GFDL01011942 JAV23103.1 CM000070 EAL26822.2 CH479186 EDW39372.1 OUUW01000041 SPP89936.1 GECU01021427 GECU01014371 GECU01009143 JAS86279.1 JAS93335.1 JAS98563.1 KA649803 KA649804 AFP64433.1 CH933806 EDW14558.1 GAMC01011209 GAMC01011208 JAB95347.1 KK852437 KDR23917.1 NEVH01019966 PNF22107.1 PNF22108.1 GEYN01000124 JAV02005.1 GBXI01008396 GBXI01007656 JAD05896.1 JAD06636.1 JRES01001112 KNC25461.1 KK853306 KDR08706.1 GAKP01015109 JAC43843.1 CH477308 EAT44022.1 GDHF01034036 GDHF01031892 GDHF01023085 JAI18278.1 JAI20422.1 JAI29229.1 GEZM01060604 JAV70980.1 JXUM01107306 JXUM01107307 JXUM01107308 JXUM01107309 KQ565130 KXJ71303.1 GEBQ01004508 JAT35469.1 GBHO01001405 GBHO01001404 JAG42199.1 JAG42200.1 CH477278 EAT45071.1 DS232338 EDS40306.1 CH916369 EDV93158.1 KQ971345 EFA05022.2 AGBW02013325 OWR43604.1 AF482940 AAM28515.1 ATLV01020821 KE525308 KFB45839.1 GDHC01021497 JAP97131.1 KQ460845 KPJ12051.1 AAAB01008859 EGK96646.1 CH954182 EDV53097.1 APCN01000618 AXCM01001395 EDS40305.1 KQ982566 KYQ54757.1 GEBQ01002690 JAT37287.1 ADMH02002143 ETN58290.1 GL888067 EGI68276.1 KQ978543 KYN30213.1 GL452916 EFN76589.1 KY172016 AUC64292.1 RSAL01000020 RVE52654.1 ABLF02025683
JX531674 AGO45859.1 HQ704866 AET36546.1 RSAL01000244 RVE43603.1 PCG75660.1 KQ460325 KPJ15745.1 KPJ15746.1 KQ459564 KPI99678.1 ODYU01002605 SOQ40327.1 JX531659 AGO45844.1 MF687531 ATJ44457.1 NWSH01000211 PCG78358.1 GEYN01000134 JAV01995.1 JX531665 AGO45850.1 AGBW02009734 OWR50018.1 ODYU01012535 SOQ58905.1 AF482926 AAM28501.1 GU952764 ADO85597.1 GEDC01030129 GEDC01013889 JAS07169.1 JAS23409.1 CH902617 EDV44111.1 LBMM01016179 KMQ84512.1 CH940652 EDW59212.1 CH964272 EDW84429.1 AJWK01009460 AJWK01009461 AF482918 AAM28493.1 GDIQ01004277 JAN90460.1 JXJN01007190 CCAG010012086 EZ422854 ADD19130.1 CM000364 EDX14986.1 CM000160 EDW99077.1 AE014297 AY047565 AAF57020.1 AAK77297.1 GECZ01031245 JAS38524.1 NNAY01004348 OXU18023.1 AF482899 AAM33369.1 GFDL01011942 JAV23103.1 CM000070 EAL26822.2 CH479186 EDW39372.1 OUUW01000041 SPP89936.1 GECU01021427 GECU01014371 GECU01009143 JAS86279.1 JAS93335.1 JAS98563.1 KA649803 KA649804 AFP64433.1 CH933806 EDW14558.1 GAMC01011209 GAMC01011208 JAB95347.1 KK852437 KDR23917.1 NEVH01019966 PNF22107.1 PNF22108.1 GEYN01000124 JAV02005.1 GBXI01008396 GBXI01007656 JAD05896.1 JAD06636.1 JRES01001112 KNC25461.1 KK853306 KDR08706.1 GAKP01015109 JAC43843.1 CH477308 EAT44022.1 GDHF01034036 GDHF01031892 GDHF01023085 JAI18278.1 JAI20422.1 JAI29229.1 GEZM01060604 JAV70980.1 JXUM01107306 JXUM01107307 JXUM01107308 JXUM01107309 KQ565130 KXJ71303.1 GEBQ01004508 JAT35469.1 GBHO01001405 GBHO01001404 JAG42199.1 JAG42200.1 CH477278 EAT45071.1 DS232338 EDS40306.1 CH916369 EDV93158.1 KQ971345 EFA05022.2 AGBW02013325 OWR43604.1 AF482940 AAM28515.1 ATLV01020821 KE525308 KFB45839.1 GDHC01021497 JAP97131.1 KQ460845 KPJ12051.1 AAAB01008859 EGK96646.1 CH954182 EDV53097.1 APCN01000618 AXCM01001395 EDS40305.1 KQ982566 KYQ54757.1 GEBQ01002690 JAT37287.1 ADMH02002143 ETN58290.1 GL888067 EGI68276.1 KQ978543 KYN30213.1 GL452916 EFN76589.1 KY172016 AUC64292.1 RSAL01000020 RVE52654.1 ABLF02025683
Proteomes
UP000005204
UP000218220
UP000283053
UP000053240
UP000053268
UP000007151
+ More
UP000007801 UP000092443 UP000078200 UP000036403 UP000192221 UP000008792 UP000007798 UP000092461 UP000091820 UP000192223 UP000092445 UP000092460 UP000092444 UP000000304 UP000002282 UP000000803 UP000215335 UP000095301 UP000001819 UP000008744 UP000095300 UP000268350 UP000009192 UP000027135 UP000235965 UP000002358 UP000037069 UP000075901 UP000008820 UP000069940 UP000249989 UP000075880 UP000075881 UP000002320 UP000079169 UP000001070 UP000007266 UP000030765 UP000007062 UP000008711 UP000075840 UP000075883 UP000075809 UP000000673 UP000007755 UP000078492 UP000008237 UP000007819
UP000007801 UP000092443 UP000078200 UP000036403 UP000192221 UP000008792 UP000007798 UP000092461 UP000091820 UP000192223 UP000092445 UP000092460 UP000092444 UP000000304 UP000002282 UP000000803 UP000215335 UP000095301 UP000001819 UP000008744 UP000095300 UP000268350 UP000009192 UP000027135 UP000235965 UP000002358 UP000037069 UP000075901 UP000008820 UP000069940 UP000249989 UP000075880 UP000075881 UP000002320 UP000079169 UP000001070 UP000007266 UP000030765 UP000007062 UP000008711 UP000075840 UP000075883 UP000075809 UP000000673 UP000007755 UP000078492 UP000008237 UP000007819
Pfam
PF00487 FA_desaturase
ProteinModelPortal
H9JYD0
G9BY58
A0A2A4JV26
T1RU52
G9BY59
A0A3S2NSR1
+ More
A0A2A4JW88 A0A194REN0 A0A194RIH1 A0A194Q2W5 A0A2H1VHN0 T1RU43 A0A291P0E4 A0A2A4K356 A0A1L8D6H7 T1RTD4 A0A212F8H4 A0A2H1X0N9 Q8MZX4 E3TMU9 A0A1B6DCJ1 B3LZD2 A0A1A9XNV7 A0A1A9VPX3 A0A0J7K2H2 A0A1W4VQC5 B4M6D6 B4NGT2 A0A1B0CEY1 A0A1A9WN17 A0A1W4X6D0 Q8MZY2 A0A1A9ZF92 A0A0P6IHM7 A0A1B0B1J8 A0A1B0GE34 D3TN52 B4R1J8 B4PLU0 Q9VA94 A0A1B6EKR5 A0A232EI48 A0A1I8N2Y4 Q8N001 A0A1Q3F6G7 Q29C24 B4GNP6 A0A1I8PAY4 A0A3B0KVP4 A0A1B6IH88 T1PM50 B4K9T3 W8B9K3 A0A067RJC1 A0A2J7Q0J6 K7J1Y0 A0A2J7Q0J2 K7IYW5 A0A1L8D6I7 A0A0A1X4V7 A0A1W4WVL1 A0A0L0BZP6 A0A067QIN8 A0A182SRI4 A0A034VQK3 Q17CJ9 A0A0K8U130 A0A1Y1LF74 A0A182H3A8 A0A182JFM4 A0A182KA51 A0A1B6MHV3 A0A1S4F5C8 A0A0A9ZCN0 Q17EY7 A0A1S4F557 B0X4F3 A0A3Q0IQ53 B4JEM7 D2A5U2 A0A212EQ61 Q8MZW0 A0A084W6J5 A0A146KPE5 A0A194R858 F5HK94 B3P7X0 A0A182HUB6 A0A182M1I7 B0X4F2 A0A151X2U1 A0A1B6MMW0 W5J5Z7 F4WBY7 A0A151JS66 E2C646 A0A2H4WB77 A0A3S2NP26 J9KA67
A0A2A4JW88 A0A194REN0 A0A194RIH1 A0A194Q2W5 A0A2H1VHN0 T1RU43 A0A291P0E4 A0A2A4K356 A0A1L8D6H7 T1RTD4 A0A212F8H4 A0A2H1X0N9 Q8MZX4 E3TMU9 A0A1B6DCJ1 B3LZD2 A0A1A9XNV7 A0A1A9VPX3 A0A0J7K2H2 A0A1W4VQC5 B4M6D6 B4NGT2 A0A1B0CEY1 A0A1A9WN17 A0A1W4X6D0 Q8MZY2 A0A1A9ZF92 A0A0P6IHM7 A0A1B0B1J8 A0A1B0GE34 D3TN52 B4R1J8 B4PLU0 Q9VA94 A0A1B6EKR5 A0A232EI48 A0A1I8N2Y4 Q8N001 A0A1Q3F6G7 Q29C24 B4GNP6 A0A1I8PAY4 A0A3B0KVP4 A0A1B6IH88 T1PM50 B4K9T3 W8B9K3 A0A067RJC1 A0A2J7Q0J6 K7J1Y0 A0A2J7Q0J2 K7IYW5 A0A1L8D6I7 A0A0A1X4V7 A0A1W4WVL1 A0A0L0BZP6 A0A067QIN8 A0A182SRI4 A0A034VQK3 Q17CJ9 A0A0K8U130 A0A1Y1LF74 A0A182H3A8 A0A182JFM4 A0A182KA51 A0A1B6MHV3 A0A1S4F5C8 A0A0A9ZCN0 Q17EY7 A0A1S4F557 B0X4F3 A0A3Q0IQ53 B4JEM7 D2A5U2 A0A212EQ61 Q8MZW0 A0A084W6J5 A0A146KPE5 A0A194R858 F5HK94 B3P7X0 A0A182HUB6 A0A182M1I7 B0X4F2 A0A151X2U1 A0A1B6MMW0 W5J5Z7 F4WBY7 A0A151JS66 E2C646 A0A2H4WB77 A0A3S2NP26 J9KA67
PDB
4YMK
E-value=3.15795e-54,
Score=533
Ontologies
PATHWAY
GO
PANTHER
Topology
SignalP
Position: 1 - 15,
Likelihood: 0.532914
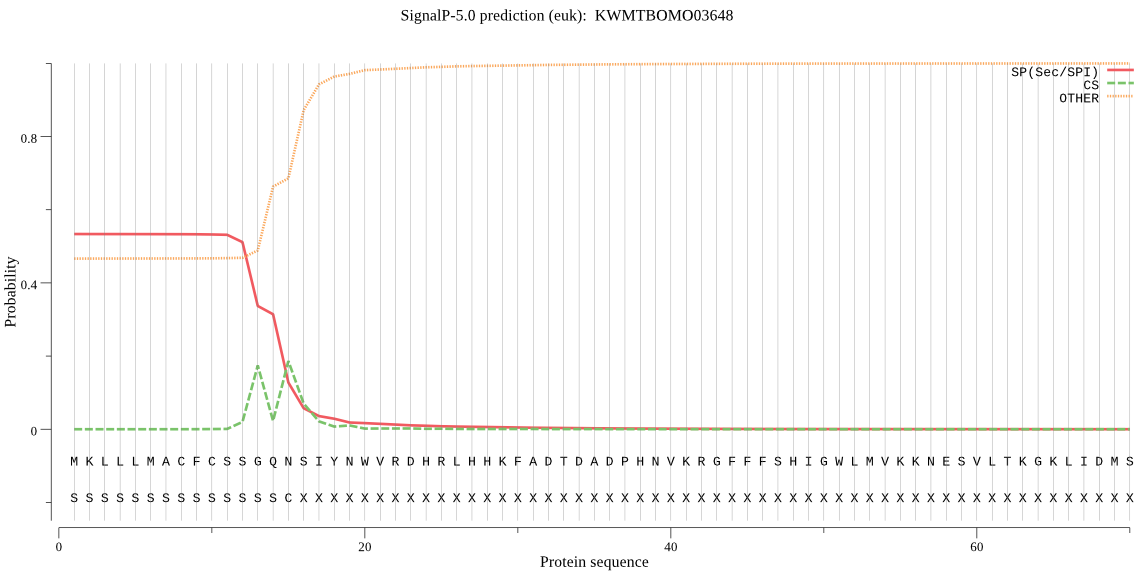
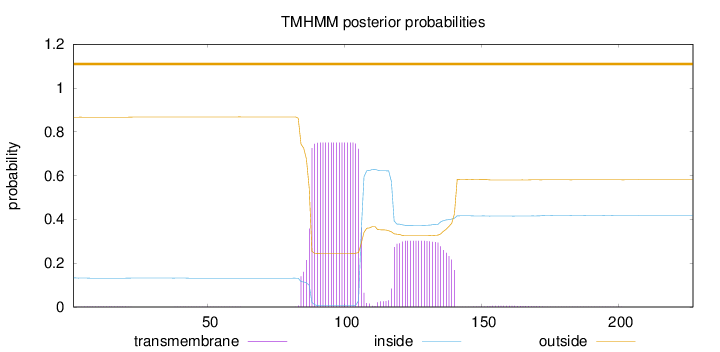
Length:
227
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
21.6363
Exp number, first 60 AAs:
0.03722
Total prob of N-in:
0.13289
outside
1 - 227
Population Genetic Test Statistics
Pi
17.595629
Theta
19.058607
Tajima's D
-1.092329
CLR
12.350829
CSRT
0.122793860306985
Interpretation
Uncertain
