Gene
KWMTBOMO03645
Annotation
PREDICTED:_RNA-directed_DNA_polymerase_from_mobile_element_jockey-like_[Amyelois_transitella]
Location in the cell
Mitochondrial Reliability : 2.409
Sequence
CDS
ATGGACATTTTGGCTTCTCTTCATTCAAAAAAAAGTTTCTCATCATTTTGGAAGAGTACTAAAAAATTTGATAAATCACCAAGTCTTCCAGTTAGCGTGGAGGGTGAGAGTGACTACATTGGAATAGCAAATATATTTAAAGATCATTTTAAAGTAATTTCTCCATTAAAGACAGCGTCGTTGAGGAATGGTGCTGAGTTCTGTCAATCAGGTTTGCCAGTTCATTTTACATGTAAAGAAATAAAAACTGTTATAAAGGGTATGAAAAGAGGCAAGTCTCCTGGTAATGATGGACTCAGCATCGAGCATCTCCAGAATTCTGGTGTTCACCTGCCGAGAGTTTTGACTATGTTGTTCAATTTGTTTGTGGGGCATTCTCACTTACCAAGGGAGATGTTGGACACAATAGTTGTACCTATTGTAAAAAACAGGACAGGAGACATAGCGGACAAGGGAAATTATAGACCAATTTCACTGGCAACCGTCATAGCCAAGGTGTTTGACAGCGTGCTTGACTCACATCTCAGTGGATACACTAAACTGCATGATACACAGTTTGGATTTTGTGCTGGGCTATCTACTGAGAGTGCCATTTTTGCACTGAAGCATGCTGTCAAATACTACACTGATAGGAAAACACCAGTGTATGCCTGTTTCCTGGACCTATCAAAAGCCTTTGACCTTGTTTCCTATAATATCTTGTGGGAAAAGCTACTGGGAGAGGGTGTACTTCCTTCATTAGTAAATATATTATCTTTGTGGTATGAGAAGCAGAGGAACTTTGTACGGTGGTCCAATGTACTGTCAGACCCTTACAAACTAGAATGTGGTGTAAGGCAGGGTGGTTTGACCTCCCCGAGGCTCTTTAATATATATGTAAATAAGCTACTTGAGGAGCTCAGTAGCACACATGTTGGTTGTCATATAGATAATTTGTGTCTTAATAACATTAGCTATGCGGATGACATGGTGCTGCTGAGCCCATCAATATGTGGGTTAAGAAAGTTGCTTTCCATATGTGAATCCTATGCTGAGGATCATGGCTTAAAGTATAACCCTAGTAAAAGTGAGCTGTTGGTGTTCAGAGGTGTAAACAAAAGTCCACCTCATGTCCCGCCTCTAATGCTCGGTGGCTTACCTGTACAAAGAGTAGCACAGTTCAAATATCTAGGGCATATTGTAACTGAGGAATTAAAAGATGATATGGACATGGAAAGGGAACGCAGGGCATTGGCTGTCAGAGGGAATGTAGTGGCACAAAGATTTGCAGGTTGTTCAGTGCCGGTCAAAATTACCCTCTTCAAGGCTTACTGCCAAACCTTTTACACAAGCGGTCTTTGGGCTACTCATACCCAAAAAACGGATAATGCCCTGCGCATACAATATAATAATGCCTTCAGGGCGTTGTTGAGGCTTCCACCTTATTGTAGTGCATCAGGCATGTTCGCCGATGCGCACACGGATGACTACTTCGCGATAATGCGCAAAAAGGTGGCATCCATTATGCGTAGAGTAGGAGGAAGCACCAACAACGTCTTGAGGGTACTAGCTGAAAGACTGGATGGCCCGACCCTGGGTAGGTTCATCAAGTTGCATGTTCTACATGCTGCTTGA
Protein
MDILASLHSKKSFSSFWKSTKKFDKSPSLPVSVEGESDYIGIANIFKDHFKVISPLKTASLRNGAEFCQSGLPVHFTCKEIKTVIKGMKRGKSPGNDGLSIEHLQNSGVHLPRVLTMLFNLFVGHSHLPREMLDTIVVPIVKNRTGDIADKGNYRPISLATVIAKVFDSVLDSHLSGYTKLHDTQFGFCAGLSTESAIFALKHAVKYYTDRKTPVYACFLDLSKAFDLVSYNILWEKLLGEGVLPSLVNILSLWYEKQRNFVRWSNVLSDPYKLECGVRQGGLTSPRLFNIYVNKLLEELSSTHVGCHIDNLCLNNISYADDMVLLSPSICGLRKLLSICESYAEDHGLKYNPSKSELLVFRGVNKSPPHVPPLMLGGLPVQRVAQFKYLGHIVTEELKDDMDMERERRALAVRGNVVAQRFAGCSVPVKITLFKAYCQTFYTSGLWATHTQKTDNALRIQYNNAFRALLRLPPYCSASGMFADAHTDDYFAIMRKKVASIMRRVGGSTNNVLRVLAERLDGPTLGRFIKLHVLHAA
Summary
Uniprot
A0A2W1BGX6
A0A2W1CP14
A0A3S2PB24
A0A2A4KA90
A0A2W1BE63
A0A2W1BJX4
+ More
A0A2W1BUS0 A0A3S2NHF1 S4NNA8 A0A2W1BC01 A0A3B3HNF2 A0A3B3I5I8 A0A3B3IJ19 A0A3P9KQQ7 A0A3B3I0P1 A0A3P8PZH5 A0A1A8UAJ7 A0A3P8RHR9 A0A146RF43 A0A146R782 A0A146RB37 A0A2H1VMX5 A0A146ZPI4 A0A146P365 A0A146RD82 A0A0P4VXS2 A0A3C1S3F5 A0A354GI36 A0A0A9YSH8 D5LB38 A0A1Y1K7T4 A0A1T2KXF3 A0A146M389 W4YPF0 W4Y5D2 A0A2S2QAF2 A0A1Y1M3I5 A0A1Y1LZ08 A0A3B1JRA2 A0A151QVZ1 A0A0A9YHJ5 A0A1B6KRX2 A0A0K8SCZ7 A0A0A9Y6A7 A0A224XIQ4 A0A2D4KEY8 A0A0J7KZE8 A0A146KVE5 A0A224X5Y1 A0A1Y1K5U2 A0A1Y1K319
A0A2W1BUS0 A0A3S2NHF1 S4NNA8 A0A2W1BC01 A0A3B3HNF2 A0A3B3I5I8 A0A3B3IJ19 A0A3P9KQQ7 A0A3B3I0P1 A0A3P8PZH5 A0A1A8UAJ7 A0A3P8RHR9 A0A146RF43 A0A146R782 A0A146RB37 A0A2H1VMX5 A0A146ZPI4 A0A146P365 A0A146RD82 A0A0P4VXS2 A0A3C1S3F5 A0A354GI36 A0A0A9YSH8 D5LB38 A0A1Y1K7T4 A0A1T2KXF3 A0A146M389 W4YPF0 W4Y5D2 A0A2S2QAF2 A0A1Y1M3I5 A0A1Y1LZ08 A0A3B1JRA2 A0A151QVZ1 A0A0A9YHJ5 A0A1B6KRX2 A0A0K8SCZ7 A0A0A9Y6A7 A0A224XIQ4 A0A2D4KEY8 A0A0J7KZE8 A0A146KVE5 A0A224X5Y1 A0A1Y1K5U2 A0A1Y1K319
Pubmed
EMBL
KZ150185
PZC72467.1
NHMN01024349
PZC87407.1
RSAL01000132
RVE46368.1
+ More
NWSH01000012 PCG80896.1 KZ150122 PZC73158.1 KZ150039 PZC74591.1 KZ149940 PZC76937.1 RSAL01004459 RVE40147.1 GAIX01014011 JAA78549.1 KZ150625 PZC70440.1 HAEJ01003895 SBS44352.1 GCES01119762 JAQ66560.1 GCES01122004 JAQ64318.1 GCES01120657 JAQ65665.1 ODYU01003417 SOQ42157.1 GCES01018090 JAR68233.1 GCES01147931 JAQ38391.1 GCES01120011 JAQ66311.1 GDRN01099830 JAI58698.1 DMQF01000507 HAO15811.1 DNUT01000366 HBI40674.1 GBHO01007597 JAG36007.1 GU815089 ADF18552.1 GEZM01094272 GEZM01094271 JAV55745.1 MPRK01000277 OOZ37539.1 GDHC01004388 JAQ14241.1 AAGJ04127121 AAGJ04127122 AAGJ04127123 AAGJ04058037 GGMS01004979 MBY74182.1 GEZM01043747 JAV78865.1 GEZM01043748 JAV78864.1 KQ484600 KYP34423.1 GBHO01011985 JAG31619.1 GEBQ01025973 JAT14004.1 GBRD01015171 JAG50655.1 GBHO01016428 JAG27176.1 GFTR01008507 JAW07919.1 IACL01060951 LAB07281.1 LBMM01001656 KMQ95932.1 GDHC01019542 JAP99086.1 GFTR01008539 JAW07887.1 GEZM01094174 JAV55841.1 GEZM01094173 JAV55843.1
NWSH01000012 PCG80896.1 KZ150122 PZC73158.1 KZ150039 PZC74591.1 KZ149940 PZC76937.1 RSAL01004459 RVE40147.1 GAIX01014011 JAA78549.1 KZ150625 PZC70440.1 HAEJ01003895 SBS44352.1 GCES01119762 JAQ66560.1 GCES01122004 JAQ64318.1 GCES01120657 JAQ65665.1 ODYU01003417 SOQ42157.1 GCES01018090 JAR68233.1 GCES01147931 JAQ38391.1 GCES01120011 JAQ66311.1 GDRN01099830 JAI58698.1 DMQF01000507 HAO15811.1 DNUT01000366 HBI40674.1 GBHO01007597 JAG36007.1 GU815089 ADF18552.1 GEZM01094272 GEZM01094271 JAV55745.1 MPRK01000277 OOZ37539.1 GDHC01004388 JAQ14241.1 AAGJ04127121 AAGJ04127122 AAGJ04127123 AAGJ04058037 GGMS01004979 MBY74182.1 GEZM01043747 JAV78865.1 GEZM01043748 JAV78864.1 KQ484600 KYP34423.1 GBHO01011985 JAG31619.1 GEBQ01025973 JAT14004.1 GBRD01015171 JAG50655.1 GBHO01016428 JAG27176.1 GFTR01008507 JAW07919.1 IACL01060951 LAB07281.1 LBMM01001656 KMQ95932.1 GDHC01019542 JAP99086.1 GFTR01008539 JAW07887.1 GEZM01094174 JAV55841.1 GEZM01094173 JAV55843.1
Proteomes
PRIDE
Interpro
Gene 3D
CDD
ProteinModelPortal
A0A2W1BGX6
A0A2W1CP14
A0A3S2PB24
A0A2A4KA90
A0A2W1BE63
A0A2W1BJX4
+ More
A0A2W1BUS0 A0A3S2NHF1 S4NNA8 A0A2W1BC01 A0A3B3HNF2 A0A3B3I5I8 A0A3B3IJ19 A0A3P9KQQ7 A0A3B3I0P1 A0A3P8PZH5 A0A1A8UAJ7 A0A3P8RHR9 A0A146RF43 A0A146R782 A0A146RB37 A0A2H1VMX5 A0A146ZPI4 A0A146P365 A0A146RD82 A0A0P4VXS2 A0A3C1S3F5 A0A354GI36 A0A0A9YSH8 D5LB38 A0A1Y1K7T4 A0A1T2KXF3 A0A146M389 W4YPF0 W4Y5D2 A0A2S2QAF2 A0A1Y1M3I5 A0A1Y1LZ08 A0A3B1JRA2 A0A151QVZ1 A0A0A9YHJ5 A0A1B6KRX2 A0A0K8SCZ7 A0A0A9Y6A7 A0A224XIQ4 A0A2D4KEY8 A0A0J7KZE8 A0A146KVE5 A0A224X5Y1 A0A1Y1K5U2 A0A1Y1K319
A0A2W1BUS0 A0A3S2NHF1 S4NNA8 A0A2W1BC01 A0A3B3HNF2 A0A3B3I5I8 A0A3B3IJ19 A0A3P9KQQ7 A0A3B3I0P1 A0A3P8PZH5 A0A1A8UAJ7 A0A3P8RHR9 A0A146RF43 A0A146R782 A0A146RB37 A0A2H1VMX5 A0A146ZPI4 A0A146P365 A0A146RD82 A0A0P4VXS2 A0A3C1S3F5 A0A354GI36 A0A0A9YSH8 D5LB38 A0A1Y1K7T4 A0A1T2KXF3 A0A146M389 W4YPF0 W4Y5D2 A0A2S2QAF2 A0A1Y1M3I5 A0A1Y1LZ08 A0A3B1JRA2 A0A151QVZ1 A0A0A9YHJ5 A0A1B6KRX2 A0A0K8SCZ7 A0A0A9Y6A7 A0A224XIQ4 A0A2D4KEY8 A0A0J7KZE8 A0A146KVE5 A0A224X5Y1 A0A1Y1K5U2 A0A1Y1K319
PDB
5IRG
E-value=7.99029e-05,
Score=111
Ontologies
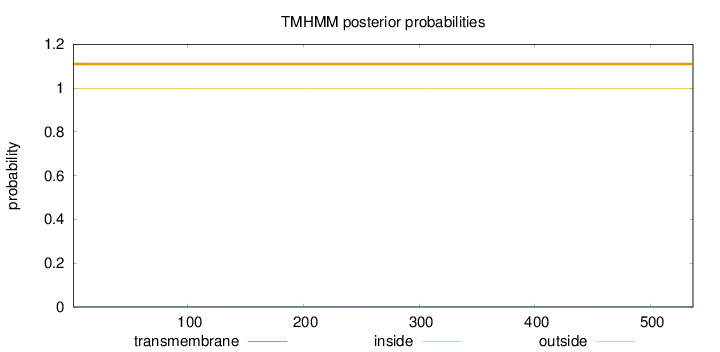
Topology
Length:
537
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02735
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00119
outside
1 - 537
Population Genetic Test Statistics
Pi
4.031435
Theta
14.2568
Tajima's D
-1.828663
CLR
14.516054
CSRT
0.0243987800609969
Interpretation
Uncertain
