Gene
KWMTBOMO03639 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA006257
Annotation
ras_protein_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 3.229
Sequence
CDS
ATGCGCGAGTACAAAATAGTTGTGTTAGGCAGCGGAGGCGTGGGAAAGTCCGCCCTGACAGTGCAATTTGTACAAGGCATCTTTGTCGAGAAATACGACCCAACCATCGAAGACAGCTACAGGAAACAAGTGGAAGTTGATGGACAGCAATGTATGCTCGAGATCCTGGATACCGCAGGTACAGAGCAGTTTACCGCGATGCGGGATCTCTACATGAAGAACGGGCAAGGGTTCGTGTTGGTGTACTCTATCACAGCACAGTCTACGTTCAATGACCTACAGGATCTACGTGAACAAATCCTGCGAGTGAAGGACACGACCGACGTGCCAATGGTTCTGGTAGGCAACAAAACCGATCTAGAGGCGGAGCGCGTGGTTGGCAAGGAACAGGGACAAAATCTGGCGCGCCACTTCAACTGCGCTTTCATGGAGACGTCGGCAAAAGCGAAAATACATGTTAACGATGTGTTCTATGACTTAGTGAGACAGATCAACAAGAAGTCCCCCAAGAAGGATGAACACAAACCTAATAAACGGAAATGTATTATTCTGTAA
Protein
MREYKIVVLGSGGVGKSALTVQFVQGIFVEKYDPTIEDSYRKQVEVDGQQCMLEILDTAGTEQFTAMRDLYMKNGQGFVLVYSITAQSTFNDLQDLREQILRVKDTTDVPMVLVGNKTDLEAERVVGKEQGQNLARHFNCAFMETSAKAKIHVNDVFYDLVRQINKKSPKKDEHKPNKRKCIIL
Summary
Uniprot
Q5KT57
A0A2H1X0W0
A0A2W1BZP5
A0A2A4KA48
A0A194R663
A0A194QFT7
+ More
A0A0K8TNP4 A0A023EII8 T1DIU4 A0A1Q3EUA6 Q16W03 A0A1B6LRZ9 A0A1B6GP06 A0A1B6HG13 A0A0L7R368 A0A2A3EB23 A0A087ZZ92 E1U2Q1 V9IDY2 A0A182YJV9 A0A182I0H0 F5HMC8 T1E8E7 A0A2M3YY38 A0A2M4A3V8 W5J2D7 A0A232EVJ8 K7IYK9 A0A1S3ID90 A0A0L7L5Y3 A0A132A3W3 A0A0C9RF02 S4P7C5 A0A3R7M6R9 A0A1B1IJM9 A0A1S3ID85 N6TII0 E0VID5 Q6IQP4 A0A1B6EED1 D0EWY2 A7SB93 A0A0P4W1Z3 R4G4H7 A0A2P2HVY9 H2SV97 H3D4L1 A0A1A8K0Y8 A0A1A8BV72 A0A1A7ZCR6 E4W4T2 A0A195CQM9 A0A195E0B7 E9ISJ6 F4X3L6 A0A195BN20 A0A195F2F7 A0A158P003 E2AYA9 A0A151X3U7 A0A026VTT4 A0A1A8EPL5 A0A3Q2YTY0 E2BY81 A0A3B3TT34 A0A3Q2G4B0 A0A1D1V6H7 A0A3B3B9J6 A0A1A8SDZ1 A0A1A8NBR2 A0A0L8H8U9 A0A0N5AMJ8 R7TFA5 A0A3R7P9I3 T1JD94 T1ILM8 A0A3P8X1R5 A0A224XQI5 A0A0V0GBE7 A0A023F9T5 A0A069DQ68 A0A3Q3KKD9 A0A0B1PLA7 A0A077Z7Y5 A0A0N5E546 A0A3B4G7S6 A0A3B5ADZ4 A0A3Q2VHG0 C3KHG6 A0A3Q0RZJ9 I3KJ34 A0A3P9DM20 A0A0B6Z9P2 C3ZAF6 W5MWG6 A0A3B4XZE1 A0A3Q1EHJ6 A0A3B5Q8X6 A0A3B4T605 A0A3P9MXR3
A0A0K8TNP4 A0A023EII8 T1DIU4 A0A1Q3EUA6 Q16W03 A0A1B6LRZ9 A0A1B6GP06 A0A1B6HG13 A0A0L7R368 A0A2A3EB23 A0A087ZZ92 E1U2Q1 V9IDY2 A0A182YJV9 A0A182I0H0 F5HMC8 T1E8E7 A0A2M3YY38 A0A2M4A3V8 W5J2D7 A0A232EVJ8 K7IYK9 A0A1S3ID90 A0A0L7L5Y3 A0A132A3W3 A0A0C9RF02 S4P7C5 A0A3R7M6R9 A0A1B1IJM9 A0A1S3ID85 N6TII0 E0VID5 Q6IQP4 A0A1B6EED1 D0EWY2 A7SB93 A0A0P4W1Z3 R4G4H7 A0A2P2HVY9 H2SV97 H3D4L1 A0A1A8K0Y8 A0A1A8BV72 A0A1A7ZCR6 E4W4T2 A0A195CQM9 A0A195E0B7 E9ISJ6 F4X3L6 A0A195BN20 A0A195F2F7 A0A158P003 E2AYA9 A0A151X3U7 A0A026VTT4 A0A1A8EPL5 A0A3Q2YTY0 E2BY81 A0A3B3TT34 A0A3Q2G4B0 A0A1D1V6H7 A0A3B3B9J6 A0A1A8SDZ1 A0A1A8NBR2 A0A0L8H8U9 A0A0N5AMJ8 R7TFA5 A0A3R7P9I3 T1JD94 T1ILM8 A0A3P8X1R5 A0A224XQI5 A0A0V0GBE7 A0A023F9T5 A0A069DQ68 A0A3Q3KKD9 A0A0B1PLA7 A0A077Z7Y5 A0A0N5E546 A0A3B4G7S6 A0A3B5ADZ4 A0A3Q2VHG0 C3KHG6 A0A3Q0RZJ9 I3KJ34 A0A3P9DM20 A0A0B6Z9P2 C3ZAF6 W5MWG6 A0A3B4XZE1 A0A3Q1EHJ6 A0A3B5Q8X6 A0A3B4T605 A0A3P9MXR3
Pubmed
19121390
19946625
28756777
26354079
26369729
24945155
+ More
24330624 17510324 25244985 12364791 14747013 17210077 20920257 23761445 28648823 20075255 26227816 26555130 23622113 27349204 23537049 20566863 23594743 17615350 27129103 21551351 15496914 21282665 21719571 21347285 20798317 24508170 30249741 27649274 29451363 23254933 24487278 25474469 26334808 25186727 18563158 23542700
24330624 17510324 25244985 12364791 14747013 17210077 20920257 23761445 28648823 20075255 26227816 26555130 23622113 27349204 23537049 20566863 23594743 17615350 27129103 21551351 15496914 21282665 21719571 21347285 20798317 24508170 30249741 27649274 29451363 23254933 24487278 25474469 26334808 25186727 18563158 23542700
EMBL
BABH01026102
AB170011
BAD83770.1
ODYU01012532
SOQ58898.1
KZ149893
+ More
PZC78915.1 NWSH01000017 PCG80768.1 KQ460647 KPJ13157.1 KQ459011 KPJ04352.1 GDAI01001591 JAI16012.1 GAPW01004838 JAC08760.1 GALA01000751 JAA94101.1 GFDL01016209 JAV18836.1 CH477578 EAT38763.1 GEBQ01013573 JAT26404.1 GECZ01005614 JAS64155.1 GECU01034167 JAS73539.1 KQ414663 KOC65278.1 KZ288295 PBC28973.1 EU365127 ACA64426.1 JR039085 JR039086 AEY58523.1 AEY58524.1 APCN01002077 AAAB01008987 EAL38518.3 EGK97449.1 EGK97450.1 GAMD01002242 JAA99348.1 GGFM01000432 MBW21183.1 GGFK01002110 MBW35431.1 ADMH02002134 GGFL01004309 ETN58407.1 MBW68487.1 NNAY01001983 OXU22382.1 JTDY01002682 KOB70938.1 JXLN01010201 KPM05305.1 GBYB01005566 JAG75333.1 GAIX01004694 JAA87866.1 QCYY01002023 ROT73516.1 KX274687 ANR94953.1 APGK01024770 KB740562 KB632399 ENN80239.1 ERL94854.1 DS235196 EEB13141.1 BX664742 BC071360 AAH71360.1 GEDC01001031 JAS36267.1 GQ903728 ACX54109.1 DS469614 EDO39047.1 GDKW01000948 JAI55647.1 ACPB03025138 GAHY01000761 JAA76749.1 IACF01000114 LAB65922.1 HAED01017054 HAEE01005973 SBR25993.1 HADZ01007626 HAEA01015028 SBP71567.1 HADY01001803 HAEJ01017861 SBP40288.1 EU871827 ACJ66625.1 KQ977394 KYN03011.1 KQ979955 KYN18541.1 GL765434 EFZ16260.1 GL888624 EGI58813.1 KQ976440 KYM86579.1 KQ981856 KYN34566.1 ADTU01005184 GL443864 EFN61568.1 KQ982557 KYQ55083.1 KK107929 QOIP01000003 EZA47112.1 RLU25111.1 HAEB01002285 SBQ48812.1 GL451420 EFN79360.1 BDGG01000002 GAU94413.1 HAEH01016823 HAEI01013400 SBS15869.1 HAEF01005544 HAEG01002068 SBR66520.1 KQ418957 KOF85180.1 AMQN01013324 KB310130 ELT92419.1 QCYY01001263 ROT79295.1 JH432102 JH430855 GFTR01003072 JAW13354.1 GECL01000683 JAP05441.1 GBBI01000557 JAC18155.1 GBGD01003127 JAC85762.1 KN538427 KHJ40867.1 HG805916 CDW54800.1 BT082381 ACQ58088.1 AERX01002589 AERX01002590 AERX01002591 AERX01002592 HACG01017555 HACG01017556 CEK64420.1 CEK64421.1 GG666602 EEN50359.1 AHAT01014319
PZC78915.1 NWSH01000017 PCG80768.1 KQ460647 KPJ13157.1 KQ459011 KPJ04352.1 GDAI01001591 JAI16012.1 GAPW01004838 JAC08760.1 GALA01000751 JAA94101.1 GFDL01016209 JAV18836.1 CH477578 EAT38763.1 GEBQ01013573 JAT26404.1 GECZ01005614 JAS64155.1 GECU01034167 JAS73539.1 KQ414663 KOC65278.1 KZ288295 PBC28973.1 EU365127 ACA64426.1 JR039085 JR039086 AEY58523.1 AEY58524.1 APCN01002077 AAAB01008987 EAL38518.3 EGK97449.1 EGK97450.1 GAMD01002242 JAA99348.1 GGFM01000432 MBW21183.1 GGFK01002110 MBW35431.1 ADMH02002134 GGFL01004309 ETN58407.1 MBW68487.1 NNAY01001983 OXU22382.1 JTDY01002682 KOB70938.1 JXLN01010201 KPM05305.1 GBYB01005566 JAG75333.1 GAIX01004694 JAA87866.1 QCYY01002023 ROT73516.1 KX274687 ANR94953.1 APGK01024770 KB740562 KB632399 ENN80239.1 ERL94854.1 DS235196 EEB13141.1 BX664742 BC071360 AAH71360.1 GEDC01001031 JAS36267.1 GQ903728 ACX54109.1 DS469614 EDO39047.1 GDKW01000948 JAI55647.1 ACPB03025138 GAHY01000761 JAA76749.1 IACF01000114 LAB65922.1 HAED01017054 HAEE01005973 SBR25993.1 HADZ01007626 HAEA01015028 SBP71567.1 HADY01001803 HAEJ01017861 SBP40288.1 EU871827 ACJ66625.1 KQ977394 KYN03011.1 KQ979955 KYN18541.1 GL765434 EFZ16260.1 GL888624 EGI58813.1 KQ976440 KYM86579.1 KQ981856 KYN34566.1 ADTU01005184 GL443864 EFN61568.1 KQ982557 KYQ55083.1 KK107929 QOIP01000003 EZA47112.1 RLU25111.1 HAEB01002285 SBQ48812.1 GL451420 EFN79360.1 BDGG01000002 GAU94413.1 HAEH01016823 HAEI01013400 SBS15869.1 HAEF01005544 HAEG01002068 SBR66520.1 KQ418957 KOF85180.1 AMQN01013324 KB310130 ELT92419.1 QCYY01001263 ROT79295.1 JH432102 JH430855 GFTR01003072 JAW13354.1 GECL01000683 JAP05441.1 GBBI01000557 JAC18155.1 GBGD01003127 JAC85762.1 KN538427 KHJ40867.1 HG805916 CDW54800.1 BT082381 ACQ58088.1 AERX01002589 AERX01002590 AERX01002591 AERX01002592 HACG01017555 HACG01017556 CEK64420.1 CEK64421.1 GG666602 EEN50359.1 AHAT01014319
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000008820
UP000053825
+ More
UP000242457 UP000005203 UP000076408 UP000075840 UP000007062 UP000000673 UP000215335 UP000002358 UP000085678 UP000037510 UP000283509 UP000019118 UP000030742 UP000009046 UP000000437 UP000001593 UP000015103 UP000005226 UP000007303 UP000078542 UP000078492 UP000007755 UP000078540 UP000078541 UP000005205 UP000000311 UP000075809 UP000053097 UP000279307 UP000264820 UP000008237 UP000261500 UP000265020 UP000186922 UP000261560 UP000053454 UP000046393 UP000014760 UP000265120 UP000261640 UP000030665 UP000046395 UP000261460 UP000261400 UP000264840 UP000261340 UP000005207 UP000265160 UP000001554 UP000018468 UP000261360 UP000257200 UP000002852 UP000261420 UP000242638
UP000242457 UP000005203 UP000076408 UP000075840 UP000007062 UP000000673 UP000215335 UP000002358 UP000085678 UP000037510 UP000283509 UP000019118 UP000030742 UP000009046 UP000000437 UP000001593 UP000015103 UP000005226 UP000007303 UP000078542 UP000078492 UP000007755 UP000078540 UP000078541 UP000005205 UP000000311 UP000075809 UP000053097 UP000279307 UP000264820 UP000008237 UP000261500 UP000265020 UP000186922 UP000261560 UP000053454 UP000046393 UP000014760 UP000265120 UP000261640 UP000030665 UP000046395 UP000261460 UP000261400 UP000264840 UP000261340 UP000005207 UP000265160 UP000001554 UP000018468 UP000261360 UP000257200 UP000002852 UP000261420 UP000242638
PRIDE
Pfam
PF00071 Ras
Interpro
SUPFAM
SSF52540
SSF52540
CDD
ProteinModelPortal
Q5KT57
A0A2H1X0W0
A0A2W1BZP5
A0A2A4KA48
A0A194R663
A0A194QFT7
+ More
A0A0K8TNP4 A0A023EII8 T1DIU4 A0A1Q3EUA6 Q16W03 A0A1B6LRZ9 A0A1B6GP06 A0A1B6HG13 A0A0L7R368 A0A2A3EB23 A0A087ZZ92 E1U2Q1 V9IDY2 A0A182YJV9 A0A182I0H0 F5HMC8 T1E8E7 A0A2M3YY38 A0A2M4A3V8 W5J2D7 A0A232EVJ8 K7IYK9 A0A1S3ID90 A0A0L7L5Y3 A0A132A3W3 A0A0C9RF02 S4P7C5 A0A3R7M6R9 A0A1B1IJM9 A0A1S3ID85 N6TII0 E0VID5 Q6IQP4 A0A1B6EED1 D0EWY2 A7SB93 A0A0P4W1Z3 R4G4H7 A0A2P2HVY9 H2SV97 H3D4L1 A0A1A8K0Y8 A0A1A8BV72 A0A1A7ZCR6 E4W4T2 A0A195CQM9 A0A195E0B7 E9ISJ6 F4X3L6 A0A195BN20 A0A195F2F7 A0A158P003 E2AYA9 A0A151X3U7 A0A026VTT4 A0A1A8EPL5 A0A3Q2YTY0 E2BY81 A0A3B3TT34 A0A3Q2G4B0 A0A1D1V6H7 A0A3B3B9J6 A0A1A8SDZ1 A0A1A8NBR2 A0A0L8H8U9 A0A0N5AMJ8 R7TFA5 A0A3R7P9I3 T1JD94 T1ILM8 A0A3P8X1R5 A0A224XQI5 A0A0V0GBE7 A0A023F9T5 A0A069DQ68 A0A3Q3KKD9 A0A0B1PLA7 A0A077Z7Y5 A0A0N5E546 A0A3B4G7S6 A0A3B5ADZ4 A0A3Q2VHG0 C3KHG6 A0A3Q0RZJ9 I3KJ34 A0A3P9DM20 A0A0B6Z9P2 C3ZAF6 W5MWG6 A0A3B4XZE1 A0A3Q1EHJ6 A0A3B5Q8X6 A0A3B4T605 A0A3P9MXR3
A0A0K8TNP4 A0A023EII8 T1DIU4 A0A1Q3EUA6 Q16W03 A0A1B6LRZ9 A0A1B6GP06 A0A1B6HG13 A0A0L7R368 A0A2A3EB23 A0A087ZZ92 E1U2Q1 V9IDY2 A0A182YJV9 A0A182I0H0 F5HMC8 T1E8E7 A0A2M3YY38 A0A2M4A3V8 W5J2D7 A0A232EVJ8 K7IYK9 A0A1S3ID90 A0A0L7L5Y3 A0A132A3W3 A0A0C9RF02 S4P7C5 A0A3R7M6R9 A0A1B1IJM9 A0A1S3ID85 N6TII0 E0VID5 Q6IQP4 A0A1B6EED1 D0EWY2 A7SB93 A0A0P4W1Z3 R4G4H7 A0A2P2HVY9 H2SV97 H3D4L1 A0A1A8K0Y8 A0A1A8BV72 A0A1A7ZCR6 E4W4T2 A0A195CQM9 A0A195E0B7 E9ISJ6 F4X3L6 A0A195BN20 A0A195F2F7 A0A158P003 E2AYA9 A0A151X3U7 A0A026VTT4 A0A1A8EPL5 A0A3Q2YTY0 E2BY81 A0A3B3TT34 A0A3Q2G4B0 A0A1D1V6H7 A0A3B3B9J6 A0A1A8SDZ1 A0A1A8NBR2 A0A0L8H8U9 A0A0N5AMJ8 R7TFA5 A0A3R7P9I3 T1JD94 T1ILM8 A0A3P8X1R5 A0A224XQI5 A0A0V0GBE7 A0A023F9T5 A0A069DQ68 A0A3Q3KKD9 A0A0B1PLA7 A0A077Z7Y5 A0A0N5E546 A0A3B4G7S6 A0A3B5ADZ4 A0A3Q2VHG0 C3KHG6 A0A3Q0RZJ9 I3KJ34 A0A3P9DM20 A0A0B6Z9P2 C3ZAF6 W5MWG6 A0A3B4XZE1 A0A3Q1EHJ6 A0A3B5Q8X6 A0A3B4T605 A0A3P9MXR3
PDB
4M8N
E-value=4.44288e-80,
Score=754
Ontologies
GO
PANTHER
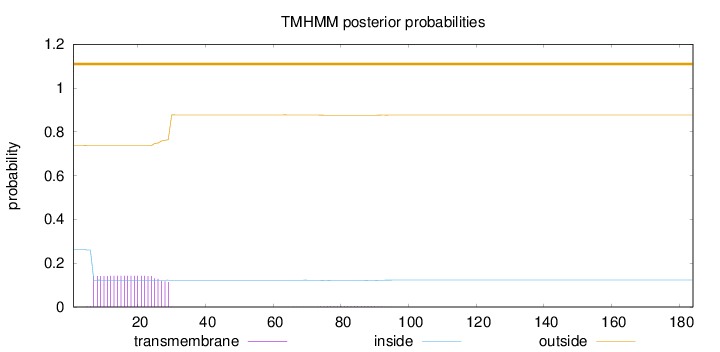
Topology
Length:
184
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
3.22247
Exp number, first 60 AAs:
3.16566
Total prob of N-in:
0.26311
outside
1 - 184
Population Genetic Test Statistics
Pi
16.067832
Theta
15.226791
Tajima's D
-0.03792
CLR
0
CSRT
0.365631718414079
Interpretation
Uncertain
