Gene
KWMTBOMO03638
Pre Gene Modal
BGIBMGA006338
Annotation
PREDICTED:_rootletin_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.701
Sequence
CDS
ATGGTTTTTAGGGGTTTCATTTATTGTCGACTACAAAACGAGGCGGAGGAGCGCGAGCGGTGCGCCAGCGAGGCGGCGGCGGCGCGGCGGCACGGCGCCGAGCTGGAGGCCGTGCTGCAGGCCGCGCGGGCCGACCTCGCCGCCGCGCGCCAGCGGGCCGCCGAGCTGGACGAGGCCTGTCGCGCCAGGGACCAGGAATTAGTGCTGCGTCTAGAAGACTGCCGCTCGAAGGAGCGTAGACTTGAAGAACTGAAGCACAACTTGGAAGTTTGTCTCGCCGATGCGACGCAGCAAATACAGGAATTGAAGGCGCGGCTCGGCGGGTGCGAGGGCCGGTGCCGCGCGCTGGAGGCGTCCCTGAGCCAGTGCGAGGGCGGCAGGCGCGAGGCCGAGTCCAAGCTGTCGTCGGTGGTGCACAGCTTGCGGCGCGTGTGCGGCGTGCAGCCCGACGGCTCGCTGCAGCCCGCCGCGCGCCGCCGCCTCGCCAGCCCCGCGCGCCGCTACAGCCCGCACCGAGGTCGTGACCATTCCGAGGATCGTAACGAGATAATCGACGTCGACCCGGAGTTGATCAAGAAGGGCGTCAGGAACCTGATGCACGATGTCTGTCAAATCGAGAGGGAGAAGGACGACTACAAGTCCCAAGTGACCGTGCTCAAGAAGCAGTTGAAGGAGGCGACGGAGCAGCAGGGCAAGGGCGAGGGCAAGCTGCAGACGGCGGCCGCCAGCCTGCGCGCGCTGCAGGACGAGCGGGCGCGCCTGCACGCCGCCTGCGGGAACAAGGACGCGCAGCTGCTCGCGCTGAACGAATCGATACAGTCGAAGACGGTCGAGATAAGCAGCTTGAGGGACAAGATAACCAGTTTAGAAGCAGCTCTAAGCTCCGTCACCGAAGAGAAAGTACAGGCGGAGAATAAAGCGGAAAGTCTGCGCGCGCAGCTGGCCGAGCGCGCACACGAAGCCGCCCAGCTGCGGGAGGCGCTGACGGCTGCCGAAGGTCGCGCGGCCCGCCTGGACGTGCGGCGCGCGCAGCTCGAGGGAGACGTGCAGCGCGCCGCCGTCGCCGCCAGGGACAGGGACCAGGCCCTCAAGAAACTGCAAGAGCGCTGCGAGGGCTACACGCGCACGATAGCGCAGCTGGAGGACCGCTGCACGTCGCTGAAGGGCACGGTGGAGCAGCTGTCGGCAGCGCTGCAGCGGGCCGCCACCGCTGAGGGCGAGCTGCGCGCCGAGCTGGCCAAGCTGCAGCGGCAGCTCGGCGAGGCGCGCGCGCAGGGACAGAGCGCCGCCGAGAAACTCAGGCACATACAGAAGAATATCACGAGTTGCGAGAACGAGCGGCGCGTGCTCACTGAGAAACTGGAGAGTGCCAAATCGGCTCTCGGCGAGTTAAAGCGGCAGAACTCGACGCTGGAAGACCAGGTCCACAGACTCACCAATCAGCTGGCAAACGTAGAAGTGCAACGATCTGGCCTAGAATCCCAACTGCGCATGACGACCTGGGACGGCAAGGAGTCCAACGACGGCGAGCTGGAGCGCGAGCTTCACGAGCTGCAGCGAGAGCGCTCCGAGCTCAAAGCGAAGAACGACGCCCTCCAGGACACGCTGAGGAAGCTGGAGGCGGACCGGAGGATGACCAGGCCCTCCGTGTTGAGGACCAAGAGCCACGACCGGACCGAGAAGAGCGTGTACTACTCGGATTTGGATTCAGGGCCAGATTCGACTAAGGACTCGTCGTCGCACTGCTACAAGGACAAGGAGGTGCCCTGCAGGGACAAGGGACTCTCCACCGAGCTGAGTCTGCTCGAGCTGGAGAACAGAGAGCTGAGGATGAAGATACGACGCCTCGAGAAGGAACTTGCTGATAAGGAGTCGGAGCTGGCGGTGGCCCGCAGCCGCTACCTGGGCGAGCTATCCGCTTCAGGCCCGGAGTGCCGCGACGACGCGGAGCAGTACCGCCGCGCCGCGCTGCAGGCTGAGCGGCTGCTGGAGGCGCGCGAGGCCAGCCACCGCCAGCAGATGCTGCGTCTCGAGAACCAGGCGGCGCAGCTCCGGGCGCAGCTGTCGGCGGAGATCCGGCGGCGCCAGGCGTACGTGTCGCGCAGCGGGCGCGCCGCCCGGGACGTGCAGCGCCTGCGGGCCGCGCTCGGGGACTCGCTGAGGACGGTGTCGCAGGATCCGGCCCTGGACTCTTACACATTGGAACATGAAGCCAGAAAGTTGGACAGCACCCTGGCTCACAGTCTACCGCCGCTCACCGACAGCTACGACTCATCAAAATAA
Protein
MVFRGFIYCRLQNEAEERERCASEAAAARRHGAELEAVLQAARADLAAARQRAAELDEACRARDQELVLRLEDCRSKERRLEELKHNLEVCLADATQQIQELKARLGGCEGRCRALEASLSQCEGGRREAESKLSSVVHSLRRVCGVQPDGSLQPAARRRLASPARRYSPHRGRDHSEDRNEIIDVDPELIKKGVRNLMHDVCQIEREKDDYKSQVTVLKKQLKEATEQQGKGEGKLQTAAASLRALQDERARLHAACGNKDAQLLALNESIQSKTVEISSLRDKITSLEAALSSVTEEKVQAENKAESLRAQLAERAHEAAQLREALTAAEGRAARLDVRRAQLEGDVQRAAVAARDRDQALKKLQERCEGYTRTIAQLEDRCTSLKGTVEQLSAALQRAATAEGELRAELAKLQRQLGEARAQGQSAAEKLRHIQKNITSCENERRVLTEKLESAKSALGELKRQNSTLEDQVHRLTNQLANVEVQRSGLESQLRMTTWDGKESNDGELERELHELQRERSELKAKNDALQDTLRKLEADRRMTRPSVLRTKSHDRTEKSVYYSDLDSGPDSTKDSSSHCYKDKEVPCRDKGLSTELSLLELENRELRMKIRRLEKELADKESELAVARSRYLGELSASGPECRDDAEQYRRAALQAERLLEAREASHRQQMLRLENQAAQLRAQLSAEIRRRQAYVSRSGRAARDVQRLRAALGDSLRTVSQDPALDSYTLEHEARKLDSTLAHSLPPLTDSYDSSK
Summary
Uniprot
H9J9Z3
A0A2W1C1S3
A0A2H1WN22
A0A194QHD9
A0A2A4K9L5
A0A2A4KAK9
+ More
A0A212FGJ2 U4UUM7 A0A2J7RAI3 A0A154PFW9 A0A1B6DR95 A0A1Y1KLC3 E2B1W1 A0A088AIK1 A0A067RCM1 A0A1Y1KP05 A0A2A3ELA4 E2BI77 E9J840 A0A151WFQ2 A0A151IGA7 A0A158NCC9 F4WWW8 E0VQE7 A0A0L7RDF9 A0A195BX65 A0A195FA33 B4HGI0 A0A3L8DJD9 A0A026VX42 A0A151JRC7 W8BFI9 A0A0M9A7A4 A0A310SIL3 K7J1S2 A0A1B0DFF2 A0A0R3NM45 A0A034W3Q4 Q8MSN3 W8B8C7 A0A0P8YHI3 B4PL19 A0A1J1HJW0 B3P794 A0A1W4V4Y4 A0A1S4FNQ4 A0A3B0JKG4 Q298K1 A0A3B0KAL6 B3M0M2 Q9VCD1 A0A0K8WED7 A0A0K8UUX9 Q16UG5 A0A0M5JCU3 B4JIG6 B4MBY9 B0W3H0 A0A0A9Y2R0 A0A146MI46 D6WAQ4 B4K7D8 A0A084WV24 A0A023F204 A0A182GEU3 A0A1S4HI61 A0A0L0CLJ9 B4NBN2 B4G420 A0A182NKX5 A0A1I8Q0E1 A0A182KFC9 A0A1I8MNC7 A0A182R837 A0A182J463 A0A1B0BJN2 A0A0J7L423 A0A182MK66 A0A182VSA1 A0A182Q9W2 A0A182F8K7 A0A1A9X182 W5JSS1 A0A1A9ZLB0 A0A1A9VR09 A0A182HJV1 A0A182XK40 A0A182YM45 Q7QDG9 A0A1B0G867 A0A182KX30 A0A182PN00 A0A0A9Y8A8 A0A232FCT5 A0A182U1F7 A0A2S2NHV3
A0A212FGJ2 U4UUM7 A0A2J7RAI3 A0A154PFW9 A0A1B6DR95 A0A1Y1KLC3 E2B1W1 A0A088AIK1 A0A067RCM1 A0A1Y1KP05 A0A2A3ELA4 E2BI77 E9J840 A0A151WFQ2 A0A151IGA7 A0A158NCC9 F4WWW8 E0VQE7 A0A0L7RDF9 A0A195BX65 A0A195FA33 B4HGI0 A0A3L8DJD9 A0A026VX42 A0A151JRC7 W8BFI9 A0A0M9A7A4 A0A310SIL3 K7J1S2 A0A1B0DFF2 A0A0R3NM45 A0A034W3Q4 Q8MSN3 W8B8C7 A0A0P8YHI3 B4PL19 A0A1J1HJW0 B3P794 A0A1W4V4Y4 A0A1S4FNQ4 A0A3B0JKG4 Q298K1 A0A3B0KAL6 B3M0M2 Q9VCD1 A0A0K8WED7 A0A0K8UUX9 Q16UG5 A0A0M5JCU3 B4JIG6 B4MBY9 B0W3H0 A0A0A9Y2R0 A0A146MI46 D6WAQ4 B4K7D8 A0A084WV24 A0A023F204 A0A182GEU3 A0A1S4HI61 A0A0L0CLJ9 B4NBN2 B4G420 A0A182NKX5 A0A1I8Q0E1 A0A182KFC9 A0A1I8MNC7 A0A182R837 A0A182J463 A0A1B0BJN2 A0A0J7L423 A0A182MK66 A0A182VSA1 A0A182Q9W2 A0A182F8K7 A0A1A9X182 W5JSS1 A0A1A9ZLB0 A0A1A9VR09 A0A182HJV1 A0A182XK40 A0A182YM45 Q7QDG9 A0A1B0G867 A0A182KX30 A0A182PN00 A0A0A9Y8A8 A0A232FCT5 A0A182U1F7 A0A2S2NHV3
Pubmed
19121390
28756777
26354079
22118469
23537049
28004739
+ More
20798317 24845553 21282665 21347285 21719571 20566863 17994087 30249741 24508170 24495485 20075255 15632085 25348373 17550304 17510324 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25401762 26823975 18362917 19820115 24438588 25474469 26483478 26108605 25315136 20920257 23761445 25244985 12364791 20966253 28648823
20798317 24845553 21282665 21347285 21719571 20566863 17994087 30249741 24508170 24495485 20075255 15632085 25348373 17550304 17510324 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25401762 26823975 18362917 19820115 24438588 25474469 26483478 26108605 25315136 20920257 23761445 25244985 12364791 20966253 28648823
EMBL
BABH01026097
BABH01026098
BABH01026099
BABH01026100
KZ149893
PZC78916.1
+ More
ODYU01009807 SOQ54470.1 KQ459011 KPJ04350.1 NWSH01000017 PCG80769.1 PCG80770.1 AGBW02008658 OWR52848.1 KB632375 ERL93850.1 NEVH01006569 PNF37841.1 KQ434896 KZC10776.1 GEDC01009119 JAS28179.1 GEZM01080365 JAV62192.1 GL444981 EFN60317.1 KK852593 KDR20670.1 GEZM01080366 JAV62191.1 KZ288222 PBC32052.1 GL448457 EFN84608.1 GL768788 EFZ11012.1 KQ983211 KYQ46667.1 KQ977728 KYN00257.1 ADTU01011816 GL888417 EGI61211.1 DS235418 EEB15603.1 KQ414614 KOC68883.1 KQ976396 KYM92885.1 KQ981727 KYN37072.1 CH480815 EDW43433.1 QOIP01000008 RLU19968.1 KK107648 EZA48327.1 KQ978602 KYN29933.1 GAMC01009088 JAB97467.1 KQ435720 KOX78690.1 KQ760869 OAD58796.1 AJVK01058717 CM000070 KRT00357.1 GAKP01010539 JAC48413.1 AY118691 AAM50551.1 GAMC01009090 JAB97465.1 CH902617 KPU80731.1 CM000160 EDW99004.2 CVRI01000006 CRK87834.1 CH954182 EDV53914.1 OUUW01000007 SPP82715.1 EAL27948.1 SPP82716.1 EDV44269.1 AE014297 AAF56238.3 AAN13982.3 AHN57496.1 GDHF01003104 JAI49210.1 GDHF01021837 JAI30477.1 CH477622 EAT38172.1 CP012526 ALC47431.1 CH916369 EDV93047.1 CH940656 EDW58610.1 DS231831 EDS31313.1 GBHO01016267 JAG27337.1 GDHC01000213 JAQ18416.1 KQ971312 EEZ97966.2 CH933806 EDW14262.1 KL646512 KFB54068.1 GBBI01003763 JAC14949.1 JXUM01058733 JXUM01058734 KQ562016 KXJ76893.1 ATLV01011398 JRES01000321 KNC32319.1 CH964232 EDW81196.1 CH479179 EDW24431.1 JXJN01015472 LBMM01000915 KMQ97229.1 AXCM01000679 AXCN02000109 ADMH02000197 ETN67402.1 APCN01002209 AAAB01008851 EAA07351.5 CCAG010000199 GBHO01016271 JAG27333.1 NNAY01000427 OXU28485.1 GGMR01004106 MBY16725.1
ODYU01009807 SOQ54470.1 KQ459011 KPJ04350.1 NWSH01000017 PCG80769.1 PCG80770.1 AGBW02008658 OWR52848.1 KB632375 ERL93850.1 NEVH01006569 PNF37841.1 KQ434896 KZC10776.1 GEDC01009119 JAS28179.1 GEZM01080365 JAV62192.1 GL444981 EFN60317.1 KK852593 KDR20670.1 GEZM01080366 JAV62191.1 KZ288222 PBC32052.1 GL448457 EFN84608.1 GL768788 EFZ11012.1 KQ983211 KYQ46667.1 KQ977728 KYN00257.1 ADTU01011816 GL888417 EGI61211.1 DS235418 EEB15603.1 KQ414614 KOC68883.1 KQ976396 KYM92885.1 KQ981727 KYN37072.1 CH480815 EDW43433.1 QOIP01000008 RLU19968.1 KK107648 EZA48327.1 KQ978602 KYN29933.1 GAMC01009088 JAB97467.1 KQ435720 KOX78690.1 KQ760869 OAD58796.1 AJVK01058717 CM000070 KRT00357.1 GAKP01010539 JAC48413.1 AY118691 AAM50551.1 GAMC01009090 JAB97465.1 CH902617 KPU80731.1 CM000160 EDW99004.2 CVRI01000006 CRK87834.1 CH954182 EDV53914.1 OUUW01000007 SPP82715.1 EAL27948.1 SPP82716.1 EDV44269.1 AE014297 AAF56238.3 AAN13982.3 AHN57496.1 GDHF01003104 JAI49210.1 GDHF01021837 JAI30477.1 CH477622 EAT38172.1 CP012526 ALC47431.1 CH916369 EDV93047.1 CH940656 EDW58610.1 DS231831 EDS31313.1 GBHO01016267 JAG27337.1 GDHC01000213 JAQ18416.1 KQ971312 EEZ97966.2 CH933806 EDW14262.1 KL646512 KFB54068.1 GBBI01003763 JAC14949.1 JXUM01058733 JXUM01058734 KQ562016 KXJ76893.1 ATLV01011398 JRES01000321 KNC32319.1 CH964232 EDW81196.1 CH479179 EDW24431.1 JXJN01015472 LBMM01000915 KMQ97229.1 AXCM01000679 AXCN02000109 ADMH02000197 ETN67402.1 APCN01002209 AAAB01008851 EAA07351.5 CCAG010000199 GBHO01016271 JAG27333.1 NNAY01000427 OXU28485.1 GGMR01004106 MBY16725.1
Proteomes
UP000005204
UP000053268
UP000218220
UP000007151
UP000030742
UP000235965
+ More
UP000076502 UP000000311 UP000005203 UP000027135 UP000242457 UP000008237 UP000075809 UP000078542 UP000005205 UP000007755 UP000009046 UP000053825 UP000078540 UP000078541 UP000001292 UP000279307 UP000053097 UP000078492 UP000053105 UP000002358 UP000092462 UP000001819 UP000007801 UP000002282 UP000183832 UP000008711 UP000192221 UP000268350 UP000000803 UP000008820 UP000092553 UP000001070 UP000008792 UP000002320 UP000007266 UP000009192 UP000030765 UP000069940 UP000249989 UP000037069 UP000007798 UP000008744 UP000075884 UP000095300 UP000075881 UP000095301 UP000075900 UP000075880 UP000092460 UP000036403 UP000075883 UP000075920 UP000075886 UP000069272 UP000091820 UP000000673 UP000092445 UP000078200 UP000075840 UP000076407 UP000076408 UP000007062 UP000092444 UP000075882 UP000075885 UP000215335 UP000075902
UP000076502 UP000000311 UP000005203 UP000027135 UP000242457 UP000008237 UP000075809 UP000078542 UP000005205 UP000007755 UP000009046 UP000053825 UP000078540 UP000078541 UP000001292 UP000279307 UP000053097 UP000078492 UP000053105 UP000002358 UP000092462 UP000001819 UP000007801 UP000002282 UP000183832 UP000008711 UP000192221 UP000268350 UP000000803 UP000008820 UP000092553 UP000001070 UP000008792 UP000002320 UP000007266 UP000009192 UP000030765 UP000069940 UP000249989 UP000037069 UP000007798 UP000008744 UP000075884 UP000095300 UP000075881 UP000095301 UP000075900 UP000075880 UP000092460 UP000036403 UP000075883 UP000075920 UP000075886 UP000069272 UP000091820 UP000000673 UP000092445 UP000078200 UP000075840 UP000076407 UP000076408 UP000007062 UP000092444 UP000075882 UP000075885 UP000215335 UP000075902
Interpro
SUPFAM
SSF53474
SSF53474
Gene 3D
ProteinModelPortal
H9J9Z3
A0A2W1C1S3
A0A2H1WN22
A0A194QHD9
A0A2A4K9L5
A0A2A4KAK9
+ More
A0A212FGJ2 U4UUM7 A0A2J7RAI3 A0A154PFW9 A0A1B6DR95 A0A1Y1KLC3 E2B1W1 A0A088AIK1 A0A067RCM1 A0A1Y1KP05 A0A2A3ELA4 E2BI77 E9J840 A0A151WFQ2 A0A151IGA7 A0A158NCC9 F4WWW8 E0VQE7 A0A0L7RDF9 A0A195BX65 A0A195FA33 B4HGI0 A0A3L8DJD9 A0A026VX42 A0A151JRC7 W8BFI9 A0A0M9A7A4 A0A310SIL3 K7J1S2 A0A1B0DFF2 A0A0R3NM45 A0A034W3Q4 Q8MSN3 W8B8C7 A0A0P8YHI3 B4PL19 A0A1J1HJW0 B3P794 A0A1W4V4Y4 A0A1S4FNQ4 A0A3B0JKG4 Q298K1 A0A3B0KAL6 B3M0M2 Q9VCD1 A0A0K8WED7 A0A0K8UUX9 Q16UG5 A0A0M5JCU3 B4JIG6 B4MBY9 B0W3H0 A0A0A9Y2R0 A0A146MI46 D6WAQ4 B4K7D8 A0A084WV24 A0A023F204 A0A182GEU3 A0A1S4HI61 A0A0L0CLJ9 B4NBN2 B4G420 A0A182NKX5 A0A1I8Q0E1 A0A182KFC9 A0A1I8MNC7 A0A182R837 A0A182J463 A0A1B0BJN2 A0A0J7L423 A0A182MK66 A0A182VSA1 A0A182Q9W2 A0A182F8K7 A0A1A9X182 W5JSS1 A0A1A9ZLB0 A0A1A9VR09 A0A182HJV1 A0A182XK40 A0A182YM45 Q7QDG9 A0A1B0G867 A0A182KX30 A0A182PN00 A0A0A9Y8A8 A0A232FCT5 A0A182U1F7 A0A2S2NHV3
A0A212FGJ2 U4UUM7 A0A2J7RAI3 A0A154PFW9 A0A1B6DR95 A0A1Y1KLC3 E2B1W1 A0A088AIK1 A0A067RCM1 A0A1Y1KP05 A0A2A3ELA4 E2BI77 E9J840 A0A151WFQ2 A0A151IGA7 A0A158NCC9 F4WWW8 E0VQE7 A0A0L7RDF9 A0A195BX65 A0A195FA33 B4HGI0 A0A3L8DJD9 A0A026VX42 A0A151JRC7 W8BFI9 A0A0M9A7A4 A0A310SIL3 K7J1S2 A0A1B0DFF2 A0A0R3NM45 A0A034W3Q4 Q8MSN3 W8B8C7 A0A0P8YHI3 B4PL19 A0A1J1HJW0 B3P794 A0A1W4V4Y4 A0A1S4FNQ4 A0A3B0JKG4 Q298K1 A0A3B0KAL6 B3M0M2 Q9VCD1 A0A0K8WED7 A0A0K8UUX9 Q16UG5 A0A0M5JCU3 B4JIG6 B4MBY9 B0W3H0 A0A0A9Y2R0 A0A146MI46 D6WAQ4 B4K7D8 A0A084WV24 A0A023F204 A0A182GEU3 A0A1S4HI61 A0A0L0CLJ9 B4NBN2 B4G420 A0A182NKX5 A0A1I8Q0E1 A0A182KFC9 A0A1I8MNC7 A0A182R837 A0A182J463 A0A1B0BJN2 A0A0J7L423 A0A182MK66 A0A182VSA1 A0A182Q9W2 A0A182F8K7 A0A1A9X182 W5JSS1 A0A1A9ZLB0 A0A1A9VR09 A0A182HJV1 A0A182XK40 A0A182YM45 Q7QDG9 A0A1B0G867 A0A182KX30 A0A182PN00 A0A0A9Y8A8 A0A232FCT5 A0A182U1F7 A0A2S2NHV3
PDB
5TBY
E-value=5.41598e-07,
Score=131
Ontologies
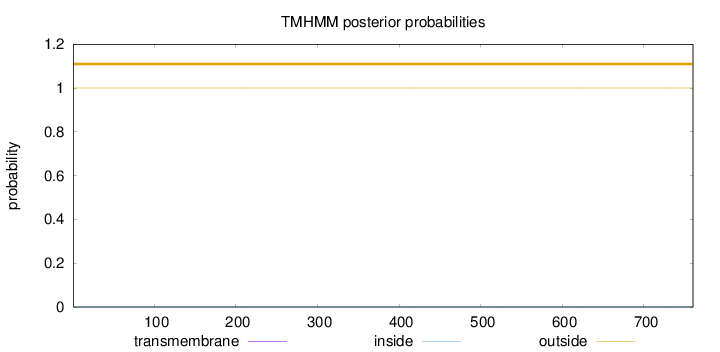
Topology
Length:
760
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00009
outside
1 - 760
Population Genetic Test Statistics
Pi
15.79138
Theta
17.293243
Tajima's D
-0.577937
CLR
0.524448
CSRT
0.224538773061347
Interpretation
Uncertain
