Gene
KWMTBOMO03636
Pre Gene Modal
BGIBMGA006337
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_rootletin-like_[Papilio_polytes]
Location in the cell
Nuclear Reliability : 3.053
Sequence
CDS
ATGGATTCTCCAGCGGGCAGTGTTGGTAGTGACTTGAGACGTCAGAATGAAGAGCTTCGTGCACGTCTTGCTGGAGAGGCGGCAGATCATAAGCGTCGTCTCGACGCGTATCGTCGTGCTCAGCAAGGCCAAGCTGCATTAGTATCGAGACTGCAAGCTAAAGTTTTGCAGTACAAGCAACGATGTGCGGAGCTCGAAAGTCACATGCTGGAGACACCCCGAAGCGAGGGTTATCGCACCAGCACGGCGCTGCAGCCGGCCGCTCTTCCCGCACCGTCGCCGCCTTCCACCGATTCGCGCAGGGATGAACGCATTAACGACTTGGAGACTGCTCTGCGTCGACTGGACGAGGAGAAACGCAAGTGTGAGAAGCTGGTAGCCCAGAATAATCTACTCCGAGAGCAACTGGAGGAGTCTCATCAGCTAAATGAAGCTCTGACAGGTGACTTGCAGAAACTGACCAATGATTGGGAGGCTCTGAGAGACGAGATGGCAGTCAAGGAGGATGAATGGAAAGATGAAGAACAGGCTTTCAATGACTACTATTCCAGCGAGCATAACCGTCTGCTAAGTCTGTGGCGCGAGGTGGTGTCTGTGAAGCGTTTCTTCTCCGAACTGCAGTCCAACACGGAACGAGATCTGTATCGGGTCAAGAACGACTTTGATTCATCTGTTAGGGAGCTTGTTGGATCATTGAGTGGATATGCTATCAATGCATTTGCAGCTCAAGGAAGTAAGGGCGAACCTCAAGCTTCAGTCGTGAGTTCGACCGCGCCAGACGTTGGAACAGTCGAAACGTTACGAGTGGAGCTCCGGGAGGTCGCCGCGCAGCGTGAGCACGCCCTGGCCGAGCTGCGGGAGCGGGACGCCCGGATACAACGGCAATAA
Protein
MDSPAGSVGSDLRRQNEELRARLAGEAADHKRRLDAYRRAQQGQAALVSRLQAKVLQYKQRCAELESHMLETPRSEGYRTSTALQPAALPAPSPPSTDSRRDERINDLETALRRLDEEKRKCEKLVAQNNLLREQLEESHQLNEALTGDLQKLTNDWEALRDEMAVKEDEWKDEEQAFNDYYSSEHNRLLSLWREVVSVKRFFSELQSNTERDLYRVKNDFDSSVRELVGSLSGYAINAFAAQGSKGEPQASVVSSTAPDVGTVETLRVELREVAAQREHALAELRERDARIQRQ
Summary
Uniprot
H9J9Z2
A0A194QHD9
A0A0L7LSY8
A0A2A4K970
A0A2W1C076
A0A2H1WXV7
+ More
A0A212FGJ2 A0A194R6L4 A0A2J7RAH3 A0A067RCM1 E2BI77 E2B1W1 A0A0J7L423 D6WAQ4 A0A3L8DJD9 A0A026VX42 N6TRB0 U4UL80 A0A154PFW9 A0A158NCC9 A0A310SIL3 A0A0L7RDF9 A0A0M9A7A4 A0A088AIK1 E9J840 A0A195BX65 A0A1J1HJW0 A0A023F204 A0A2A3ELA4 A0A3Q0IXU3 A0A151IGA7 A0A195FA33 A0A1A9XI19 A0A151WFQ2 A0A1B0BJN2 A0A1B0G867 A0A1A9ZLB0 A0A1A9VR09 F4WWW8 A0A1Y1KLB0 A0A232FCT5 T1IC61 K7J1S2 A0A1A9X182 E0VQE7 B4NBN2 B4G420 B4HGH9 A0A1I8Q0E1 Q298K1 A0A0M5JCU3 B4JIG6 A0A151JRC7 A0A0K8WED7 A0A0K8UUX9 A0A146MI46 A0A1Y1KP05 B4MBY9 A0A0A1XK31 A0A0L0CLJ9 A0A1I8MNC7 B4K7D8 A0A3B0JKG4 A0A3B0KAL6 A0A2S2Q572 J9K2K6 A0A2R7WHD7 W8B8C7 V9KZQ6 V9K942 A0A1S3HLZ9 A0A1S3HNJ8 A0A1S3P477 A0A1S3JC29
A0A212FGJ2 A0A194R6L4 A0A2J7RAH3 A0A067RCM1 E2BI77 E2B1W1 A0A0J7L423 D6WAQ4 A0A3L8DJD9 A0A026VX42 N6TRB0 U4UL80 A0A154PFW9 A0A158NCC9 A0A310SIL3 A0A0L7RDF9 A0A0M9A7A4 A0A088AIK1 E9J840 A0A195BX65 A0A1J1HJW0 A0A023F204 A0A2A3ELA4 A0A3Q0IXU3 A0A151IGA7 A0A195FA33 A0A1A9XI19 A0A151WFQ2 A0A1B0BJN2 A0A1B0G867 A0A1A9ZLB0 A0A1A9VR09 F4WWW8 A0A1Y1KLB0 A0A232FCT5 T1IC61 K7J1S2 A0A1A9X182 E0VQE7 B4NBN2 B4G420 B4HGH9 A0A1I8Q0E1 Q298K1 A0A0M5JCU3 B4JIG6 A0A151JRC7 A0A0K8WED7 A0A0K8UUX9 A0A146MI46 A0A1Y1KP05 B4MBY9 A0A0A1XK31 A0A0L0CLJ9 A0A1I8MNC7 B4K7D8 A0A3B0JKG4 A0A3B0KAL6 A0A2S2Q572 J9K2K6 A0A2R7WHD7 W8B8C7 V9KZQ6 V9K942 A0A1S3HLZ9 A0A1S3HNJ8 A0A1S3P477 A0A1S3JC29
Pubmed
EMBL
BABH01026096
BABH01026097
KQ459011
KPJ04350.1
JTDY01000164
KOB78494.1
+ More
NWSH01000017 PCG80771.1 KZ149893 PZC78917.1 ODYU01011899 SOQ57905.1 AGBW02008658 OWR52848.1 KQ460647 KPJ13159.1 NEVH01006569 PNF37836.1 KK852593 KDR20670.1 GL448457 EFN84608.1 GL444981 EFN60317.1 LBMM01000915 KMQ97229.1 KQ971312 EEZ97966.2 QOIP01000008 RLU19968.1 KK107648 EZA48327.1 APGK01023338 APGK01023339 APGK01023340 APGK01023341 APGK01023342 KB740484 ENN80568.1 KB632375 ERL93852.1 KQ434896 KZC10776.1 ADTU01011816 KQ760869 OAD58796.1 KQ414614 KOC68883.1 KQ435720 KOX78690.1 GL768788 EFZ11012.1 KQ976396 KYM92885.1 CVRI01000006 CRK87834.1 GBBI01003763 JAC14949.1 KZ288222 PBC32052.1 KQ977728 KYN00257.1 KQ981727 KYN37072.1 KQ983211 KYQ46667.1 JXJN01015472 CCAG010000199 GL888417 EGI61211.1 GEZM01080365 JAV62193.1 NNAY01000427 OXU28485.1 ACPB03006184 DS235418 EEB15603.1 CH964232 EDW81196.1 CH479179 EDW24431.1 CH480815 EDW43432.1 CM000070 EAL27948.1 CP012526 ALC47431.1 CH916369 EDV93047.1 KQ978602 KYN29933.1 GDHF01003104 JAI49210.1 GDHF01021837 JAI30477.1 GDHC01000213 JAQ18416.1 GEZM01080366 JAV62191.1 CH940656 EDW58610.1 GBXI01002558 JAD11734.1 JRES01000321 KNC32319.1 CH933806 EDW14262.1 OUUW01000007 SPP82715.1 SPP82716.1 GGMS01003538 MBY72741.1 ABLF02033859 KK854827 PTY19062.1 GAMC01009090 JAB97465.1 JW871463 AFP03981.1 JW861739 AFO94256.1
NWSH01000017 PCG80771.1 KZ149893 PZC78917.1 ODYU01011899 SOQ57905.1 AGBW02008658 OWR52848.1 KQ460647 KPJ13159.1 NEVH01006569 PNF37836.1 KK852593 KDR20670.1 GL448457 EFN84608.1 GL444981 EFN60317.1 LBMM01000915 KMQ97229.1 KQ971312 EEZ97966.2 QOIP01000008 RLU19968.1 KK107648 EZA48327.1 APGK01023338 APGK01023339 APGK01023340 APGK01023341 APGK01023342 KB740484 ENN80568.1 KB632375 ERL93852.1 KQ434896 KZC10776.1 ADTU01011816 KQ760869 OAD58796.1 KQ414614 KOC68883.1 KQ435720 KOX78690.1 GL768788 EFZ11012.1 KQ976396 KYM92885.1 CVRI01000006 CRK87834.1 GBBI01003763 JAC14949.1 KZ288222 PBC32052.1 KQ977728 KYN00257.1 KQ981727 KYN37072.1 KQ983211 KYQ46667.1 JXJN01015472 CCAG010000199 GL888417 EGI61211.1 GEZM01080365 JAV62193.1 NNAY01000427 OXU28485.1 ACPB03006184 DS235418 EEB15603.1 CH964232 EDW81196.1 CH479179 EDW24431.1 CH480815 EDW43432.1 CM000070 EAL27948.1 CP012526 ALC47431.1 CH916369 EDV93047.1 KQ978602 KYN29933.1 GDHF01003104 JAI49210.1 GDHF01021837 JAI30477.1 GDHC01000213 JAQ18416.1 GEZM01080366 JAV62191.1 CH940656 EDW58610.1 GBXI01002558 JAD11734.1 JRES01000321 KNC32319.1 CH933806 EDW14262.1 OUUW01000007 SPP82715.1 SPP82716.1 GGMS01003538 MBY72741.1 ABLF02033859 KK854827 PTY19062.1 GAMC01009090 JAB97465.1 JW871463 AFP03981.1 JW861739 AFO94256.1
Proteomes
UP000005204
UP000053268
UP000037510
UP000218220
UP000007151
UP000053240
+ More
UP000235965 UP000027135 UP000008237 UP000000311 UP000036403 UP000007266 UP000279307 UP000053097 UP000019118 UP000030742 UP000076502 UP000005205 UP000053825 UP000053105 UP000005203 UP000078540 UP000183832 UP000242457 UP000079169 UP000078542 UP000078541 UP000092443 UP000075809 UP000092460 UP000092444 UP000092445 UP000078200 UP000007755 UP000215335 UP000015103 UP000002358 UP000091820 UP000009046 UP000007798 UP000008744 UP000001292 UP000095300 UP000001819 UP000092553 UP000001070 UP000078492 UP000008792 UP000037069 UP000095301 UP000009192 UP000268350 UP000007819 UP000085678 UP000087266
UP000235965 UP000027135 UP000008237 UP000000311 UP000036403 UP000007266 UP000279307 UP000053097 UP000019118 UP000030742 UP000076502 UP000005205 UP000053825 UP000053105 UP000005203 UP000078540 UP000183832 UP000242457 UP000079169 UP000078542 UP000078541 UP000092443 UP000075809 UP000092460 UP000092444 UP000092445 UP000078200 UP000007755 UP000215335 UP000015103 UP000002358 UP000091820 UP000009046 UP000007798 UP000008744 UP000001292 UP000095300 UP000001819 UP000092553 UP000001070 UP000078492 UP000008792 UP000037069 UP000095301 UP000009192 UP000268350 UP000007819 UP000085678 UP000087266
PRIDE
Interpro
IPR026733
Rootletin
ProteinModelPortal
H9J9Z2
A0A194QHD9
A0A0L7LSY8
A0A2A4K970
A0A2W1C076
A0A2H1WXV7
+ More
A0A212FGJ2 A0A194R6L4 A0A2J7RAH3 A0A067RCM1 E2BI77 E2B1W1 A0A0J7L423 D6WAQ4 A0A3L8DJD9 A0A026VX42 N6TRB0 U4UL80 A0A154PFW9 A0A158NCC9 A0A310SIL3 A0A0L7RDF9 A0A0M9A7A4 A0A088AIK1 E9J840 A0A195BX65 A0A1J1HJW0 A0A023F204 A0A2A3ELA4 A0A3Q0IXU3 A0A151IGA7 A0A195FA33 A0A1A9XI19 A0A151WFQ2 A0A1B0BJN2 A0A1B0G867 A0A1A9ZLB0 A0A1A9VR09 F4WWW8 A0A1Y1KLB0 A0A232FCT5 T1IC61 K7J1S2 A0A1A9X182 E0VQE7 B4NBN2 B4G420 B4HGH9 A0A1I8Q0E1 Q298K1 A0A0M5JCU3 B4JIG6 A0A151JRC7 A0A0K8WED7 A0A0K8UUX9 A0A146MI46 A0A1Y1KP05 B4MBY9 A0A0A1XK31 A0A0L0CLJ9 A0A1I8MNC7 B4K7D8 A0A3B0JKG4 A0A3B0KAL6 A0A2S2Q572 J9K2K6 A0A2R7WHD7 W8B8C7 V9KZQ6 V9K942 A0A1S3HLZ9 A0A1S3HNJ8 A0A1S3P477 A0A1S3JC29
A0A212FGJ2 A0A194R6L4 A0A2J7RAH3 A0A067RCM1 E2BI77 E2B1W1 A0A0J7L423 D6WAQ4 A0A3L8DJD9 A0A026VX42 N6TRB0 U4UL80 A0A154PFW9 A0A158NCC9 A0A310SIL3 A0A0L7RDF9 A0A0M9A7A4 A0A088AIK1 E9J840 A0A195BX65 A0A1J1HJW0 A0A023F204 A0A2A3ELA4 A0A3Q0IXU3 A0A151IGA7 A0A195FA33 A0A1A9XI19 A0A151WFQ2 A0A1B0BJN2 A0A1B0G867 A0A1A9ZLB0 A0A1A9VR09 F4WWW8 A0A1Y1KLB0 A0A232FCT5 T1IC61 K7J1S2 A0A1A9X182 E0VQE7 B4NBN2 B4G420 B4HGH9 A0A1I8Q0E1 Q298K1 A0A0M5JCU3 B4JIG6 A0A151JRC7 A0A0K8WED7 A0A0K8UUX9 A0A146MI46 A0A1Y1KP05 B4MBY9 A0A0A1XK31 A0A0L0CLJ9 A0A1I8MNC7 B4K7D8 A0A3B0JKG4 A0A3B0KAL6 A0A2S2Q572 J9K2K6 A0A2R7WHD7 W8B8C7 V9KZQ6 V9K942 A0A1S3HLZ9 A0A1S3HNJ8 A0A1S3P477 A0A1S3JC29
Ontologies
KEGG
PANTHER
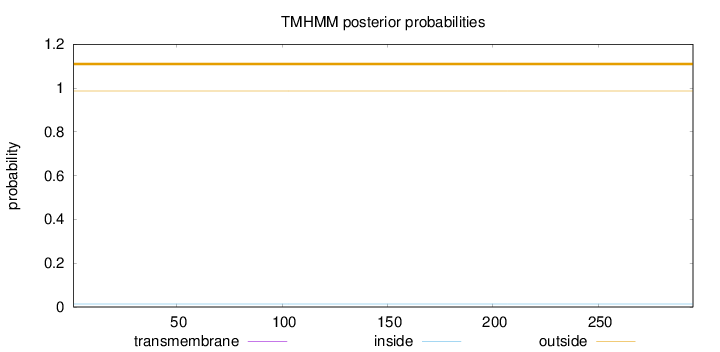
Topology
Length:
295
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00114
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01386
outside
1 - 295
Population Genetic Test Statistics
Pi
19.4707
Theta
23.028085
Tajima's D
-1.058543
CLR
0.682171
CSRT
0.127143642817859
Interpretation
Uncertain
