Pre Gene Modal
BGIBMGA006333
Annotation
PREDICTED:_ER_membrane_protein_complex_subunit_1_[Amyelois_transitella]
Full name
DNA-directed RNA polymerase subunit beta
Location in the cell
Mitochondrial Reliability : 1.158 PlasmaMembrane Reliability : 1.49
Sequence
CDS
ATGACAATAAATAATAATGTTAATTACAATCAATCCTACGTGGTGGTGGCGACGTGCGTGGACGGCGTATCGGGCGGCGTGCTGAGCGCGCACGTGCACCGGCGCGCGCGCCGCCCCGCGCTCCCGCTGCACTCCGAGCACTTCCTCGCCTACCTCTACTACAACGACAAGCAGCGCCGCCACGAGATCGCCACCCTGGAGCTGTACGAGGGCCCGGAGCGCTGGGTGCCGGCGGGCGCGGCGTTCAGCTCGCTGCGGGGGGCGCGCGCGCCGCACGTGCAGCGCCACGCCTACATCGTGCCCGCCGCGCCCGCCGCCGCCGCGCTCTCCGCCACCGAGCGCTCGCTCACCGACAAGCACGTGCTGCTGGCGCTGAGCACGGGCGCGATCGTGGAGCTGCCGTGGGCGCTGCTGGAGCCCCGGCGCGCGCTGTCCCCGGCGGGCGAGGCGCGCGAGGAGGGCCTGGTGCCCTACGCGCCCGAGCTGCCGCTGCCCGCCGAGGCCGTCGTCAACTACAACGCGACGCTGCTCCGCCTCACCCGCATACACACCGCGCCCTCCGGACTCGAGTCCACCAGCCTCGTGCTCGCCACCGGACTGGACCTGTTCTACACGAGGATAGCTCCGTCAAAGACCTTCGACCTTCTCAAGGACGACTTCGACTACTACCTCATCAGCGTAGTGCTCGCCGCGCTGGTGCTCGCCACGTACAGCACCAAGTACTTCGCTGCGAGGAAGATGCTAAAGATGGCCTGGAAGTGA
Protein
MTINNNVNYNQSYVVVATCVDGVSGGVLSAHVHRRARRPALPLHSEHFLAYLYYNDKQRRHEIATLELYEGPERWVPAGAAFSSLRGARAPHVQRHAYIVPAAPAAAALSATERSLTDKHVLLALSTGAIVELPWALLEPRRALSPAGEAREEGLVPYAPELPLPAEAVVNYNATLLRLTRIHTAPSGLESTSLVLATGLDLFYTRIAPSKTFDLLKDDFDYYLISVVLAALVLATYSTKYFAARKMLKMAWK
Summary
Description
DNA-dependent RNA polymerase catalyzes the transcription of DNA into RNA using the four ribonucleoside triphosphates as substrates.
Catalytic Activity
a ribonucleoside 5'-triphosphate + RNA(n) = diphosphate + RNA(n+1)
Similarity
Belongs to the RNA polymerase beta chain family.
Uniprot
H9J9Y8
A0A3S2P514
A0A2W1BV10
A0A2H1W3B7
A0A194QLM5
A0A0L7LQE4
+ More
A0A212F160 W8BBF4 W8BGF1 A0A0A1WRR8 A0A0K8UGD2 A0A034VDT6 A0A0K8UWL7 A0A0K8VNH1 A0A034VEG2 W8B4F5 W8BRX2 B4N8D1 A0A1A9WFN0 T1HEQ0 A0A0Q9X016 B4K7I9 B4JI35 A0A1B0BY71 Q95TQ6 A0A1B6DQ56 A0A1B6CDR2 B4M0U9 F0JAM8 A0A0C9QT61 A0A3B0KJW6 A0A1I8PCX5 A0A1I8PCW9 A0A1I8PCT7 A0A2J7PZI0 A0A1I8PCM2 A0A1B0D303 A0A0P9AQU3 B3LXF8 Q294L3 B4GMY0 I5APS5 B4R0B0 B4I4Y5 A0A1B6G4V2 A0A0B4LGV1 Q9VHY6 B3NYS9 A0A1W4VLW5 A0A1W4VZ61 A0A1B6KS38 B4PRD5 A0A1B6LTB9 A0A224XJP5 A0A0L0CKJ4 A0A1L8D938 A0A1L8D9I9 A0A1L8D9B0 A0A1L8D943 A0A0K8T908 A0A0K8TA52 A0A1I8MY05 T1PCK1 A0A146LUI5 A0A0A9WW35 A0A146LR23 A0A0A9YLC5 U5ER11 A0A336LDR2 K7J0N0 A0A154NWQ5 A0A2M3Z3W4 A0A2M3Z3V8 Q17EK4 A0A1S4F5Q7 A0A2M4ABB0 W5JK68 A0A2M4AAP1 A0A2M4AAV8 A0A0M4ERN6 A0A2M4BCP9 A0A194R6L9 A0A2A3ERP5 A0A088AG65 A0A0L7QLM8 A0A182FPR1 A0A1Q3FAS1 A0A182H0U9 A0A2R7VZ97 A0A0J7L1G5 A0A1Q3FB81 A0A1Q3FAA5 B0X5B3 A0A336LFU6 A0A1Q3FAM0 A0A310S4S8 A0A084VTM6 A0A151J8V7 A0A151JW05 A0A195CNE0
A0A212F160 W8BBF4 W8BGF1 A0A0A1WRR8 A0A0K8UGD2 A0A034VDT6 A0A0K8UWL7 A0A0K8VNH1 A0A034VEG2 W8B4F5 W8BRX2 B4N8D1 A0A1A9WFN0 T1HEQ0 A0A0Q9X016 B4K7I9 B4JI35 A0A1B0BY71 Q95TQ6 A0A1B6DQ56 A0A1B6CDR2 B4M0U9 F0JAM8 A0A0C9QT61 A0A3B0KJW6 A0A1I8PCX5 A0A1I8PCW9 A0A1I8PCT7 A0A2J7PZI0 A0A1I8PCM2 A0A1B0D303 A0A0P9AQU3 B3LXF8 Q294L3 B4GMY0 I5APS5 B4R0B0 B4I4Y5 A0A1B6G4V2 A0A0B4LGV1 Q9VHY6 B3NYS9 A0A1W4VLW5 A0A1W4VZ61 A0A1B6KS38 B4PRD5 A0A1B6LTB9 A0A224XJP5 A0A0L0CKJ4 A0A1L8D938 A0A1L8D9I9 A0A1L8D9B0 A0A1L8D943 A0A0K8T908 A0A0K8TA52 A0A1I8MY05 T1PCK1 A0A146LUI5 A0A0A9WW35 A0A146LR23 A0A0A9YLC5 U5ER11 A0A336LDR2 K7J0N0 A0A154NWQ5 A0A2M3Z3W4 A0A2M3Z3V8 Q17EK4 A0A1S4F5Q7 A0A2M4ABB0 W5JK68 A0A2M4AAP1 A0A2M4AAV8 A0A0M4ERN6 A0A2M4BCP9 A0A194R6L9 A0A2A3ERP5 A0A088AG65 A0A0L7QLM8 A0A182FPR1 A0A1Q3FAS1 A0A182H0U9 A0A2R7VZ97 A0A0J7L1G5 A0A1Q3FB81 A0A1Q3FAA5 B0X5B3 A0A336LFU6 A0A1Q3FAM0 A0A310S4S8 A0A084VTM6 A0A151J8V7 A0A151JW05 A0A195CNE0
EC Number
2.7.7.6
Pubmed
EMBL
BABH01026085
BABH01026086
RSAL01000394
RVE41980.1
KZ149893
PZC78922.1
+ More
ODYU01006057 SOQ47579.1 KQ459011 KPJ04346.1 JTDY01000336 KOB77645.1 AGBW02010943 OWR47480.1 GAMC01010548 JAB96007.1 GAMC01010547 JAB96008.1 GBXI01013006 GBXI01008352 JAD01286.1 JAD05940.1 GDHF01026939 GDHF01009119 JAI25375.1 JAI43195.1 GAKP01019003 GAKP01018997 JAC39955.1 GDHF01021328 JAI30986.1 GDHF01011887 JAI40427.1 GAKP01019001 GAKP01018999 GAKP01018996 GAKP01018994 JAC39953.1 GAMC01010550 JAB96005.1 GAMC01010549 JAB96006.1 CH964232 EDW81382.1 ACPB03020163 ACPB03020164 CH933806 KRG01519.1 EDW15333.1 CH916369 EDV92916.1 JXJN01022510 AY058611 AAL13840.1 GEDC01009484 JAS27814.1 GEDC01025712 JAS11586.1 CH940650 EDW68408.2 BT126070 ADY17769.1 GBYB01003802 JAG73569.1 OUUW01000008 SPP84058.1 NEVH01020334 PNF21748.1 AJVK01010809 AJVK01010810 CH902617 KPU79929.1 EDV42802.1 CM000070 EAL28951.2 CH479185 EDW38204.1 EIM52960.1 CM000364 EDX12035.1 CH480821 EDW55278.1 GECZ01012358 GECZ01011298 JAS57411.1 JAS58471.1 AE014297 AHN57215.1 AHN57216.1 BT031325 AAF54161.1 ABY21738.1 AHN57214.1 CH954181 EDV48192.1 GEBQ01025737 JAT14240.1 CM000160 EDW96323.2 GEBQ01013046 JAT26931.1 GFTR01008197 JAW08229.1 JRES01000266 KNC32791.1 GFDF01011111 JAV02973.1 GFDF01011110 JAV02974.1 GFDF01011109 JAV02975.1 GFDF01011112 JAV02972.1 GBRD01003822 JAG61999.1 GBRD01003821 JAG62000.1 KA646419 AFP61048.1 GDHC01008579 JAQ10050.1 GBHO01031998 JAG11606.1 GDHC01009423 JAQ09206.1 GBHO01009737 JAG33867.1 GANO01003963 JAB55908.1 UFQS01003369 UFQT01003369 SSX15564.1 SSX34928.1 KQ434773 KZC03992.1 GGFM01002449 MBW23200.1 GGFM01002455 MBW23206.1 CH477282 EAT44875.1 GGFK01004577 MBW37898.1 ADMH02001205 ETN63708.1 GGFK01004499 MBW37820.1 GGFK01004569 MBW37890.1 CP012526 ALC47864.1 GGFJ01001683 MBW50824.1 KQ460647 KPJ13164.1 KZ288197 PBC33711.1 KQ414914 KOC59525.1 GFDL01010433 JAV24612.1 JXUM01101993 JXUM01101994 KQ564691 KXJ71894.1 KK854187 PTY12792.1 LBMM01001275 KMQ96597.1 GFDL01010292 JAV24753.1 GFDL01010578 JAV24467.1 DS232374 EDS40798.1 UFQS01003368 UFQT01003368 SSX15563.1 SSX34927.1 GFDL01010445 JAV24600.1 KQ775195 OAD52265.1 ATLV01016362 KE525079 KFB41320.1 KQ979480 KYN21266.1 KQ981701 KYN37481.1 KQ977513 KYN02180.1
ODYU01006057 SOQ47579.1 KQ459011 KPJ04346.1 JTDY01000336 KOB77645.1 AGBW02010943 OWR47480.1 GAMC01010548 JAB96007.1 GAMC01010547 JAB96008.1 GBXI01013006 GBXI01008352 JAD01286.1 JAD05940.1 GDHF01026939 GDHF01009119 JAI25375.1 JAI43195.1 GAKP01019003 GAKP01018997 JAC39955.1 GDHF01021328 JAI30986.1 GDHF01011887 JAI40427.1 GAKP01019001 GAKP01018999 GAKP01018996 GAKP01018994 JAC39953.1 GAMC01010550 JAB96005.1 GAMC01010549 JAB96006.1 CH964232 EDW81382.1 ACPB03020163 ACPB03020164 CH933806 KRG01519.1 EDW15333.1 CH916369 EDV92916.1 JXJN01022510 AY058611 AAL13840.1 GEDC01009484 JAS27814.1 GEDC01025712 JAS11586.1 CH940650 EDW68408.2 BT126070 ADY17769.1 GBYB01003802 JAG73569.1 OUUW01000008 SPP84058.1 NEVH01020334 PNF21748.1 AJVK01010809 AJVK01010810 CH902617 KPU79929.1 EDV42802.1 CM000070 EAL28951.2 CH479185 EDW38204.1 EIM52960.1 CM000364 EDX12035.1 CH480821 EDW55278.1 GECZ01012358 GECZ01011298 JAS57411.1 JAS58471.1 AE014297 AHN57215.1 AHN57216.1 BT031325 AAF54161.1 ABY21738.1 AHN57214.1 CH954181 EDV48192.1 GEBQ01025737 JAT14240.1 CM000160 EDW96323.2 GEBQ01013046 JAT26931.1 GFTR01008197 JAW08229.1 JRES01000266 KNC32791.1 GFDF01011111 JAV02973.1 GFDF01011110 JAV02974.1 GFDF01011109 JAV02975.1 GFDF01011112 JAV02972.1 GBRD01003822 JAG61999.1 GBRD01003821 JAG62000.1 KA646419 AFP61048.1 GDHC01008579 JAQ10050.1 GBHO01031998 JAG11606.1 GDHC01009423 JAQ09206.1 GBHO01009737 JAG33867.1 GANO01003963 JAB55908.1 UFQS01003369 UFQT01003369 SSX15564.1 SSX34928.1 KQ434773 KZC03992.1 GGFM01002449 MBW23200.1 GGFM01002455 MBW23206.1 CH477282 EAT44875.1 GGFK01004577 MBW37898.1 ADMH02001205 ETN63708.1 GGFK01004499 MBW37820.1 GGFK01004569 MBW37890.1 CP012526 ALC47864.1 GGFJ01001683 MBW50824.1 KQ460647 KPJ13164.1 KZ288197 PBC33711.1 KQ414914 KOC59525.1 GFDL01010433 JAV24612.1 JXUM01101993 JXUM01101994 KQ564691 KXJ71894.1 KK854187 PTY12792.1 LBMM01001275 KMQ96597.1 GFDL01010292 JAV24753.1 GFDL01010578 JAV24467.1 DS232374 EDS40798.1 UFQS01003368 UFQT01003368 SSX15563.1 SSX34927.1 GFDL01010445 JAV24600.1 KQ775195 OAD52265.1 ATLV01016362 KE525079 KFB41320.1 KQ979480 KYN21266.1 KQ981701 KYN37481.1 KQ977513 KYN02180.1
Proteomes
UP000005204
UP000283053
UP000053268
UP000037510
UP000007151
UP000007798
+ More
UP000091820 UP000015103 UP000009192 UP000001070 UP000092460 UP000008792 UP000268350 UP000095300 UP000235965 UP000092462 UP000007801 UP000001819 UP000008744 UP000000304 UP000001292 UP000000803 UP000008711 UP000192221 UP000002282 UP000037069 UP000095301 UP000002358 UP000076502 UP000008820 UP000000673 UP000092553 UP000053240 UP000242457 UP000005203 UP000053825 UP000069272 UP000069940 UP000249989 UP000036403 UP000002320 UP000030765 UP000078492 UP000078541 UP000078542
UP000091820 UP000015103 UP000009192 UP000001070 UP000092460 UP000008792 UP000268350 UP000095300 UP000235965 UP000092462 UP000007801 UP000001819 UP000008744 UP000000304 UP000001292 UP000000803 UP000008711 UP000192221 UP000002282 UP000037069 UP000095301 UP000002358 UP000076502 UP000008820 UP000000673 UP000092553 UP000053240 UP000242457 UP000005203 UP000053825 UP000069272 UP000069940 UP000249989 UP000036403 UP000002320 UP000030765 UP000078492 UP000078541 UP000078542
Pfam
Interpro
IPR026895
EMC1
+ More
IPR011047 Quinoprotein_ADH-like_supfam
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR011678 EMC1_C
IPR002372 PQQ_repeat
IPR018391 PQQ_beta_propeller_repeat
IPR007121 RNA_pol_bsu_CS
IPR015712 DNA-dir_RNA_pol_su2
IPR007647 RNA_pol_Rpb2_5
IPR007642 RNA_pol_Rpb2_2
IPR014724 RNA_pol_RPB2_OB-fold
IPR037034 RNA_pol_Rpb2_2_sf
IPR007644 RNA_pol_bsu_protrusion
IPR037033 DNA-dir_RNAP_su2_hyb_sf
IPR007120 DNA-dir_RNAP_su2_dom
IPR007641 RNA_pol_Rpb2_7
IPR007645 RNA_pol_Rpb2_3
IPR007646 RNA_pol_Rpb2_4
IPR011047 Quinoprotein_ADH-like_supfam
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR011678 EMC1_C
IPR002372 PQQ_repeat
IPR018391 PQQ_beta_propeller_repeat
IPR007121 RNA_pol_bsu_CS
IPR015712 DNA-dir_RNA_pol_su2
IPR007647 RNA_pol_Rpb2_5
IPR007642 RNA_pol_Rpb2_2
IPR014724 RNA_pol_RPB2_OB-fold
IPR037034 RNA_pol_Rpb2_2_sf
IPR007644 RNA_pol_bsu_protrusion
IPR037033 DNA-dir_RNAP_su2_hyb_sf
IPR007120 DNA-dir_RNAP_su2_dom
IPR007641 RNA_pol_Rpb2_7
IPR007645 RNA_pol_Rpb2_3
IPR007646 RNA_pol_Rpb2_4
SUPFAM
SSF50998
SSF50998
ProteinModelPortal
H9J9Y8
A0A3S2P514
A0A2W1BV10
A0A2H1W3B7
A0A194QLM5
A0A0L7LQE4
+ More
A0A212F160 W8BBF4 W8BGF1 A0A0A1WRR8 A0A0K8UGD2 A0A034VDT6 A0A0K8UWL7 A0A0K8VNH1 A0A034VEG2 W8B4F5 W8BRX2 B4N8D1 A0A1A9WFN0 T1HEQ0 A0A0Q9X016 B4K7I9 B4JI35 A0A1B0BY71 Q95TQ6 A0A1B6DQ56 A0A1B6CDR2 B4M0U9 F0JAM8 A0A0C9QT61 A0A3B0KJW6 A0A1I8PCX5 A0A1I8PCW9 A0A1I8PCT7 A0A2J7PZI0 A0A1I8PCM2 A0A1B0D303 A0A0P9AQU3 B3LXF8 Q294L3 B4GMY0 I5APS5 B4R0B0 B4I4Y5 A0A1B6G4V2 A0A0B4LGV1 Q9VHY6 B3NYS9 A0A1W4VLW5 A0A1W4VZ61 A0A1B6KS38 B4PRD5 A0A1B6LTB9 A0A224XJP5 A0A0L0CKJ4 A0A1L8D938 A0A1L8D9I9 A0A1L8D9B0 A0A1L8D943 A0A0K8T908 A0A0K8TA52 A0A1I8MY05 T1PCK1 A0A146LUI5 A0A0A9WW35 A0A146LR23 A0A0A9YLC5 U5ER11 A0A336LDR2 K7J0N0 A0A154NWQ5 A0A2M3Z3W4 A0A2M3Z3V8 Q17EK4 A0A1S4F5Q7 A0A2M4ABB0 W5JK68 A0A2M4AAP1 A0A2M4AAV8 A0A0M4ERN6 A0A2M4BCP9 A0A194R6L9 A0A2A3ERP5 A0A088AG65 A0A0L7QLM8 A0A182FPR1 A0A1Q3FAS1 A0A182H0U9 A0A2R7VZ97 A0A0J7L1G5 A0A1Q3FB81 A0A1Q3FAA5 B0X5B3 A0A336LFU6 A0A1Q3FAM0 A0A310S4S8 A0A084VTM6 A0A151J8V7 A0A151JW05 A0A195CNE0
A0A212F160 W8BBF4 W8BGF1 A0A0A1WRR8 A0A0K8UGD2 A0A034VDT6 A0A0K8UWL7 A0A0K8VNH1 A0A034VEG2 W8B4F5 W8BRX2 B4N8D1 A0A1A9WFN0 T1HEQ0 A0A0Q9X016 B4K7I9 B4JI35 A0A1B0BY71 Q95TQ6 A0A1B6DQ56 A0A1B6CDR2 B4M0U9 F0JAM8 A0A0C9QT61 A0A3B0KJW6 A0A1I8PCX5 A0A1I8PCW9 A0A1I8PCT7 A0A2J7PZI0 A0A1I8PCM2 A0A1B0D303 A0A0P9AQU3 B3LXF8 Q294L3 B4GMY0 I5APS5 B4R0B0 B4I4Y5 A0A1B6G4V2 A0A0B4LGV1 Q9VHY6 B3NYS9 A0A1W4VLW5 A0A1W4VZ61 A0A1B6KS38 B4PRD5 A0A1B6LTB9 A0A224XJP5 A0A0L0CKJ4 A0A1L8D938 A0A1L8D9I9 A0A1L8D9B0 A0A1L8D943 A0A0K8T908 A0A0K8TA52 A0A1I8MY05 T1PCK1 A0A146LUI5 A0A0A9WW35 A0A146LR23 A0A0A9YLC5 U5ER11 A0A336LDR2 K7J0N0 A0A154NWQ5 A0A2M3Z3W4 A0A2M3Z3V8 Q17EK4 A0A1S4F5Q7 A0A2M4ABB0 W5JK68 A0A2M4AAP1 A0A2M4AAV8 A0A0M4ERN6 A0A2M4BCP9 A0A194R6L9 A0A2A3ERP5 A0A088AG65 A0A0L7QLM8 A0A182FPR1 A0A1Q3FAS1 A0A182H0U9 A0A2R7VZ97 A0A0J7L1G5 A0A1Q3FB81 A0A1Q3FAA5 B0X5B3 A0A336LFU6 A0A1Q3FAM0 A0A310S4S8 A0A084VTM6 A0A151J8V7 A0A151JW05 A0A195CNE0
Ontologies
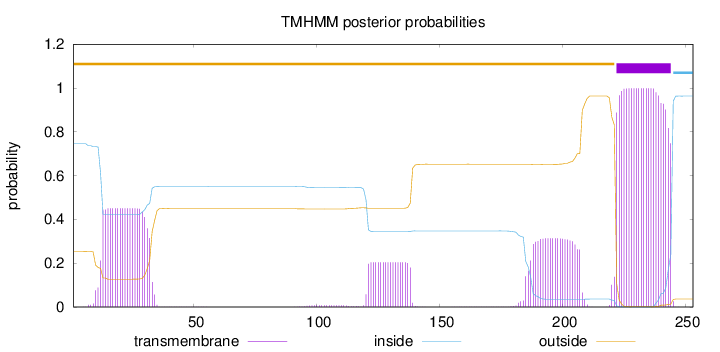
Topology
Length:
253
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
42.33975
Exp number, first 60 AAs:
9.31684999999999
Total prob of N-in:
0.74551
outside
1 - 221
TMhelix
222 - 244
inside
245 - 253
Population Genetic Test Statistics
Pi
19.611227
Theta
20.846141
Tajima's D
-0.725176
CLR
0.320485
CSRT
0.193440327983601
Interpretation
Uncertain
