Gene
KWMTBOMO03626 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA006263
Annotation
PREDICTED:_T-complex_protein_1_subunit_alpha_[Amyelois_transitella]
Full name
T-complex protein 1 subunit alpha
Alternative Name
CCT-alpha
Chaperonin containing TCP1 subunit 1
Chaperonin containing TCP1 subunit 1
Location in the cell
Cytoplasmic Reliability : 3.79
Sequence
CDS
ATGTCGACTATAGCTGCACCTTTGTCTGTAGCAGGGACAAGATCCTGCGGAGACCCAGTGAGAACACAAAATGTAATGGCAGCAGCGGCTATAGCAAACATAGTAAAGAGCTCCCTTGGGCCAGTTGGTCTGGATAAGATGCTGGTTGACGATATTGGGGATGTTACAGTCACTAACGATGGTGCCACAATATTAAAAATGCTGGAAGTGGAGCATCCTGCTGCCAAAGTCTTAGTGGAACTAGCACAGCTACAAGATGAAGAAGTTGGAGATGGTACCACATCTGTAGTCATCATTGCTGCTGAACTACTTAAGAATGCAGATGAACTAGTGAAGACCAAGATCCATCCAACAAGCATTATCTCAGGATACCGTCTCGCCTGTAAAGAAGCAGTCAAGTACATTCAGGATAACCTCACAGTAACAGTAGAGTCACTTGGCAGGCCGTCTCTGATCAACACAGCCAAAACAACCATGTCATCTAAACTAATTGGAGCTGATGCAGATTTCTTCTCGGAGATGGTTGTAGATGCTGCACAGGCAATCAAAACAATGAACCCCAGAGGAAATACTATATACCCTATCAAAGCTATTAACATTCTGAAAGCCCACGGCCGGAGTGCAAGGGAAAGTGTACTTGTCAAAGGCTATGCTCTGAACTGCACTGTAGCATCACAGGCAATGCCGAAGAAGATTCTCAATGCAAAGATCGCCTGCCTTGATTTTTCTCTGCAAAAAACTAAGATGAAATTGGGTGTTCAGGTTCTTGTGACTGATCCAGAAAAATTAGAAGCGATCCGTGCAAGAGAACTGGACATTACCAAAGAACGCTTACAGAAGATTCTCGCTACCGGCGTCAATGTTATTCTTTGCACCGGAGGCATTGATGATCTATGTTTGAAATATTTCGTTGAAGCTGGAGCAATGGGTGTCAGGAGGTGTAAGAAGGCAGATCTAAAAAGGATTGCCAAGGCTACTGGAGCTACTTTTTTGACTTCTTTGACTAATATGGATGGTGAGGAAGTATTTGAAGCTAATATGATTGGTGAAGCCGCTGAAGTGATCCAGGAACAAATCTGTGATGATCAACTTATTTTAATCAAAGGGCCGGCAGCGCGCACGGCCGCGTCGCTTATCCTGCGCGGTCCCACGGACGCGTACTGCGACGAGATGGAGCGGTCCGCGCACGACGCGCTGTGCGCCGTGCGCCGCGTGCTGGAGTCCGGCAAGCTCGTGCCCGGCGGCGGCGCGGTCGAAGCGGCACTCTCCATCTACTTGGAGAACTTTGCAACTACCCTCAGCTCCCGCGAGCAGTTGGCTATTGCAGCGTTTGCGCAGTCTCTCCTCGTGATCCCGAAGACGTTGGCGGTTAACGCGGCGCGGGACGCCACCGACCTCGTCGCCAAGCTACGAGCCTACCACAACTCATCACAAACCAAGGTGGAACACGCAAACCTCAAATGGGTAGGTCTGGATCTAACAGAAGGAACACTCCGGGACAACCTCGCAGCCGGAGTCCTCGAGCCAGCGATATCCAAGATCAAATCGTTGAAATTCGCAACAGAAGCTGCGATTACTATTCTCCGTATTGACGATATGATCAAACTCGACCCCGAACAAAGAGGCAAGAGCTATGAAGACGCTTGCAATGCCGGTGAACTGGACTAA
Protein
MSTIAAPLSVAGTRSCGDPVRTQNVMAAAAIANIVKSSLGPVGLDKMLVDDIGDVTVTNDGATILKMLEVEHPAAKVLVELAQLQDEEVGDGTTSVVIIAAELLKNADELVKTKIHPTSIISGYRLACKEAVKYIQDNLTVTVESLGRPSLINTAKTTMSSKLIGADADFFSEMVVDAAQAIKTMNPRGNTIYPIKAINILKAHGRSARESVLVKGYALNCTVASQAMPKKILNAKIACLDFSLQKTKMKLGVQVLVTDPEKLEAIRARELDITKERLQKILATGVNVILCTGGIDDLCLKYFVEAGAMGVRRCKKADLKRIAKATGATFLTSLTNMDGEEVFEANMIGEAAEVIQEQICDDQLILIKGPAARTAASLILRGPTDAYCDEMERSAHDALCAVRRVLESGKLVPGGGAVEAALSIYLENFATTLSSREQLAIAAFAQSLLVIPKTLAVNAARDATDLVAKLRAYHNSSQTKVEHANLKWVGLDLTEGTLRDNLAAGVLEPAISKIKSLKFATEAAITILRIDDMIKLDPEQRGKSYEDACNAGELD
Summary
Subunit
Heterooligomeric complex of about 850 to 900 kDa that forms two stacked rings, 12 to 16 nm in diameter.
Similarity
Belongs to the TCP-1 chaperonin family.
Keywords
ATP-binding
Chaperone
Complete proteome
Cytoplasm
Nucleotide-binding
Reference proteome
Feature
chain T-complex protein 1 subunit alpha
Uniprot
H9J9S0
A0A2H1X176
A0A2A4J635
A0A2W1BZQ1
A0A194R7F5
A0A194QLM0
+ More
A0A1Q3F3Z1 A0A1Y1MHZ9 A0A1B6C7B4 Q16W74 A0A023ETW5 A0A1J1J805 A0A1S4FLY4 B0WAH6 D6W9B9 A0A182QWL6 A0A182M9J5 A0A2J7QHG0 A0A182PD72 A0A182ND00 A0A182RV59 A0A182IZ98 A0A182V664 A0A182L730 A0A182XG44 Q7QB14 A0A182ID90 W8BY94 A0A182JPK1 A0A182SBM9 A0A0T6B8U0 A0A1B6G8J9 A0A182TLV2 V5RFY2 A0A084VBH0 A0A0L0BY42 A0A1I8Q4Q0 A0A182W1A2 A0A034WEB5 Q4AE76 A0A0K8WH30 A0A0A1WVM5 V5IAP2 Q298V2 B4G4Q0 A0A3B0KVS9 A0A2M3Z592 T1PFQ5 A0A232END1 K7J033 A0A182FPT9 T1DFU8 U5EWB8 A0A2M4BIZ6 A0A2M4A846 W5JPB5 A0A1W4WP68 B3LZZ2 J3JW40 A0A2A3E1J8 A0A088AVZ9 B4NJT3 B4MB83 B3P8P1 A0A1W4W414 A0A1L8DUD6 B4R030 A4V391 B4HEA1 P12613 B4PMA7 B4KC16 B4JSB6 A0A1A9WGN5 A0A1S3DVC9 A0A0T6AZW5 A0A1B0B2L3 A0A1A9YSY7 A0A067R174 A0A0K8TS69 A0A182G585 A0A336M5Y4 A0A158NJ24 A0A026WPB8 S4PEJ7 A0A1A9ZZ30 A0A212F181 D3TLP9 A0A1A9VFV7 E0VDT3 A0A182Y5D7 A0A1B6HYR1 A0A2J7QHG1 E9GKZ7 A0A0P5W1F5 A0A0P4ZAW1 A0A0P5TY06 A0A0M4ERP9 A0A0P5TRD2 A0A0P6FRS9
A0A1Q3F3Z1 A0A1Y1MHZ9 A0A1B6C7B4 Q16W74 A0A023ETW5 A0A1J1J805 A0A1S4FLY4 B0WAH6 D6W9B9 A0A182QWL6 A0A182M9J5 A0A2J7QHG0 A0A182PD72 A0A182ND00 A0A182RV59 A0A182IZ98 A0A182V664 A0A182L730 A0A182XG44 Q7QB14 A0A182ID90 W8BY94 A0A182JPK1 A0A182SBM9 A0A0T6B8U0 A0A1B6G8J9 A0A182TLV2 V5RFY2 A0A084VBH0 A0A0L0BY42 A0A1I8Q4Q0 A0A182W1A2 A0A034WEB5 Q4AE76 A0A0K8WH30 A0A0A1WVM5 V5IAP2 Q298V2 B4G4Q0 A0A3B0KVS9 A0A2M3Z592 T1PFQ5 A0A232END1 K7J033 A0A182FPT9 T1DFU8 U5EWB8 A0A2M4BIZ6 A0A2M4A846 W5JPB5 A0A1W4WP68 B3LZZ2 J3JW40 A0A2A3E1J8 A0A088AVZ9 B4NJT3 B4MB83 B3P8P1 A0A1W4W414 A0A1L8DUD6 B4R030 A4V391 B4HEA1 P12613 B4PMA7 B4KC16 B4JSB6 A0A1A9WGN5 A0A1S3DVC9 A0A0T6AZW5 A0A1B0B2L3 A0A1A9YSY7 A0A067R174 A0A0K8TS69 A0A182G585 A0A336M5Y4 A0A158NJ24 A0A026WPB8 S4PEJ7 A0A1A9ZZ30 A0A212F181 D3TLP9 A0A1A9VFV7 E0VDT3 A0A182Y5D7 A0A1B6HYR1 A0A2J7QHG1 E9GKZ7 A0A0P5W1F5 A0A0P4ZAW1 A0A0P5TY06 A0A0M4ERP9 A0A0P5TRD2 A0A0P6FRS9
Pubmed
19121390
28756777
26354079
28004739
17510324
24945155
+ More
26483478 18362917 19820115 20966253 12364791 14747013 17210077 24495485 24438588 26108605 25348373 16184765 25830018 15632085 17994087 25315136 28648823 20075255 20920257 23761445 22516182 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 3146529 12537569 17550304 24845553 26369729 21347285 24508170 23622113 22118469 20353571 20566863 25244985 21292972
26483478 18362917 19820115 20966253 12364791 14747013 17210077 24495485 24438588 26108605 25348373 16184765 25830018 15632085 17994087 25315136 28648823 20075255 20920257 23761445 22516182 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 3146529 12537569 17550304 24845553 26369729 21347285 24508170 23622113 22118469 20353571 20566863 25244985 21292972
EMBL
BABH01026063
ODYU01012664
SOQ59095.1
NWSH01003141
PCG66872.1
KZ149893
+ More
PZC78925.1 KQ460647 KPJ13170.1 KQ459011 KPJ04341.1 GFDL01012759 JAV22286.1 GEZM01032423 JAV84588.1 GEDC01027930 JAS09368.1 CH477573 EAT38836.1 JXUM01058052 JXUM01058053 GAPW01000783 KQ561984 JAC12815.1 KXJ76987.1 CVRI01000073 CRL07614.1 DS231872 EDS41399.1 KQ971312 EEZ98476.1 AXCN02001566 AXCM01001325 NEVH01013976 PNF28020.1 AAAB01008880 EAA08611.4 APCN01005054 GAMC01002228 JAC04328.1 LJIG01009109 KRT83738.1 GECZ01011030 JAS58739.1 KF621132 AHB33468.1 ATLV01006810 KE524386 KFB35314.1 JRES01001160 KNC24958.1 GAKP01005041 JAC53911.1 AB194475 BAE17115.1 GDHF01001955 JAI50359.1 GBXI01011616 JAD02676.1 GALX01000840 JAB67626.1 CM000070 EAL27853.1 CH479179 EDW24566.1 OUUW01000013 SPP88118.1 GGFM01002931 MBW23682.1 KA647586 AFP62215.1 NNAY01003191 OXU19832.1 GAMD01003021 JAA98569.1 GANO01001490 JAB58381.1 GGFJ01003898 MBW53039.1 GGFK01003652 MBW36973.1 ADMH02000944 ETN64609.1 CH902617 EDV44182.1 BT127458 AEE62420.1 KZ288451 PBC25575.1 CH964272 EDW85045.1 CH940656 EDW58354.1 CH954182 EDV54136.1 GFDF01004044 JAV10040.1 CM000364 EDX13936.1 AE014297 AAN13906.1 CH480815 EDW43198.1 M21159 AY118416 CM000160 EDW98012.1 CH933806 EDW16889.1 CH916373 EDV94656.1 LJIG01016411 KRT80681.1 JXJN01007683 KK852785 KDR16554.1 GDAI01000404 JAI17199.1 JXUM01042913 KQ561343 KXJ78896.1 UFQS01000609 UFQT01000609 SSX05331.1 SSX25692.1 ADTU01017356 KK107145 EZA57501.1 GAIX01004472 JAA88088.1 AGBW02010943 OWR47487.1 CCAG010014864 EZ422351 ADD18627.1 DS235088 EEB11539.1 GECU01027893 JAS79813.1 PNF28021.1 GL732550 EFX79881.1 GDIP01131700 GDIP01092293 GDIP01071546 GDIP01043251 GDIQ01077549 GDIQ01054014 LRGB01002190 JAM11422.1 JAN17188.1 KZS08616.1 GDIP01219432 JAJ03970.1 GDIP01122385 JAL81329.1 CP012526 ALC47235.1 GDIP01123844 JAL79870.1 GDIQ01046322 JAN48415.1
PZC78925.1 KQ460647 KPJ13170.1 KQ459011 KPJ04341.1 GFDL01012759 JAV22286.1 GEZM01032423 JAV84588.1 GEDC01027930 JAS09368.1 CH477573 EAT38836.1 JXUM01058052 JXUM01058053 GAPW01000783 KQ561984 JAC12815.1 KXJ76987.1 CVRI01000073 CRL07614.1 DS231872 EDS41399.1 KQ971312 EEZ98476.1 AXCN02001566 AXCM01001325 NEVH01013976 PNF28020.1 AAAB01008880 EAA08611.4 APCN01005054 GAMC01002228 JAC04328.1 LJIG01009109 KRT83738.1 GECZ01011030 JAS58739.1 KF621132 AHB33468.1 ATLV01006810 KE524386 KFB35314.1 JRES01001160 KNC24958.1 GAKP01005041 JAC53911.1 AB194475 BAE17115.1 GDHF01001955 JAI50359.1 GBXI01011616 JAD02676.1 GALX01000840 JAB67626.1 CM000070 EAL27853.1 CH479179 EDW24566.1 OUUW01000013 SPP88118.1 GGFM01002931 MBW23682.1 KA647586 AFP62215.1 NNAY01003191 OXU19832.1 GAMD01003021 JAA98569.1 GANO01001490 JAB58381.1 GGFJ01003898 MBW53039.1 GGFK01003652 MBW36973.1 ADMH02000944 ETN64609.1 CH902617 EDV44182.1 BT127458 AEE62420.1 KZ288451 PBC25575.1 CH964272 EDW85045.1 CH940656 EDW58354.1 CH954182 EDV54136.1 GFDF01004044 JAV10040.1 CM000364 EDX13936.1 AE014297 AAN13906.1 CH480815 EDW43198.1 M21159 AY118416 CM000160 EDW98012.1 CH933806 EDW16889.1 CH916373 EDV94656.1 LJIG01016411 KRT80681.1 JXJN01007683 KK852785 KDR16554.1 GDAI01000404 JAI17199.1 JXUM01042913 KQ561343 KXJ78896.1 UFQS01000609 UFQT01000609 SSX05331.1 SSX25692.1 ADTU01017356 KK107145 EZA57501.1 GAIX01004472 JAA88088.1 AGBW02010943 OWR47487.1 CCAG010014864 EZ422351 ADD18627.1 DS235088 EEB11539.1 GECU01027893 JAS79813.1 PNF28021.1 GL732550 EFX79881.1 GDIP01131700 GDIP01092293 GDIP01071546 GDIP01043251 GDIQ01077549 GDIQ01054014 LRGB01002190 JAM11422.1 JAN17188.1 KZS08616.1 GDIP01219432 JAJ03970.1 GDIP01122385 JAL81329.1 CP012526 ALC47235.1 GDIP01123844 JAL79870.1 GDIQ01046322 JAN48415.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000008820
UP000069940
+ More
UP000249989 UP000183832 UP000002320 UP000007266 UP000075886 UP000075883 UP000235965 UP000075885 UP000075884 UP000075900 UP000075880 UP000075903 UP000075882 UP000076407 UP000007062 UP000075840 UP000075881 UP000075901 UP000075902 UP000030765 UP000037069 UP000095300 UP000075920 UP000001819 UP000008744 UP000268350 UP000095301 UP000215335 UP000002358 UP000069272 UP000000673 UP000192223 UP000007801 UP000242457 UP000005203 UP000007798 UP000008792 UP000008711 UP000192221 UP000000304 UP000000803 UP000001292 UP000002282 UP000009192 UP000001070 UP000091820 UP000079169 UP000092460 UP000092443 UP000027135 UP000005205 UP000053097 UP000092445 UP000007151 UP000092444 UP000078200 UP000009046 UP000076408 UP000000305 UP000076858 UP000092553
UP000249989 UP000183832 UP000002320 UP000007266 UP000075886 UP000075883 UP000235965 UP000075885 UP000075884 UP000075900 UP000075880 UP000075903 UP000075882 UP000076407 UP000007062 UP000075840 UP000075881 UP000075901 UP000075902 UP000030765 UP000037069 UP000095300 UP000075920 UP000001819 UP000008744 UP000268350 UP000095301 UP000215335 UP000002358 UP000069272 UP000000673 UP000192223 UP000007801 UP000242457 UP000005203 UP000007798 UP000008792 UP000008711 UP000192221 UP000000304 UP000000803 UP000001292 UP000002282 UP000009192 UP000001070 UP000091820 UP000079169 UP000092460 UP000092443 UP000027135 UP000005205 UP000053097 UP000092445 UP000007151 UP000092444 UP000078200 UP000009046 UP000076408 UP000000305 UP000076858 UP000092553
PRIDE
Pfam
PF00118 Cpn60_TCP1
Interpro
Gene 3D
ProteinModelPortal
H9J9S0
A0A2H1X176
A0A2A4J635
A0A2W1BZQ1
A0A194R7F5
A0A194QLM0
+ More
A0A1Q3F3Z1 A0A1Y1MHZ9 A0A1B6C7B4 Q16W74 A0A023ETW5 A0A1J1J805 A0A1S4FLY4 B0WAH6 D6W9B9 A0A182QWL6 A0A182M9J5 A0A2J7QHG0 A0A182PD72 A0A182ND00 A0A182RV59 A0A182IZ98 A0A182V664 A0A182L730 A0A182XG44 Q7QB14 A0A182ID90 W8BY94 A0A182JPK1 A0A182SBM9 A0A0T6B8U0 A0A1B6G8J9 A0A182TLV2 V5RFY2 A0A084VBH0 A0A0L0BY42 A0A1I8Q4Q0 A0A182W1A2 A0A034WEB5 Q4AE76 A0A0K8WH30 A0A0A1WVM5 V5IAP2 Q298V2 B4G4Q0 A0A3B0KVS9 A0A2M3Z592 T1PFQ5 A0A232END1 K7J033 A0A182FPT9 T1DFU8 U5EWB8 A0A2M4BIZ6 A0A2M4A846 W5JPB5 A0A1W4WP68 B3LZZ2 J3JW40 A0A2A3E1J8 A0A088AVZ9 B4NJT3 B4MB83 B3P8P1 A0A1W4W414 A0A1L8DUD6 B4R030 A4V391 B4HEA1 P12613 B4PMA7 B4KC16 B4JSB6 A0A1A9WGN5 A0A1S3DVC9 A0A0T6AZW5 A0A1B0B2L3 A0A1A9YSY7 A0A067R174 A0A0K8TS69 A0A182G585 A0A336M5Y4 A0A158NJ24 A0A026WPB8 S4PEJ7 A0A1A9ZZ30 A0A212F181 D3TLP9 A0A1A9VFV7 E0VDT3 A0A182Y5D7 A0A1B6HYR1 A0A2J7QHG1 E9GKZ7 A0A0P5W1F5 A0A0P4ZAW1 A0A0P5TY06 A0A0M4ERP9 A0A0P5TRD2 A0A0P6FRS9
A0A1Q3F3Z1 A0A1Y1MHZ9 A0A1B6C7B4 Q16W74 A0A023ETW5 A0A1J1J805 A0A1S4FLY4 B0WAH6 D6W9B9 A0A182QWL6 A0A182M9J5 A0A2J7QHG0 A0A182PD72 A0A182ND00 A0A182RV59 A0A182IZ98 A0A182V664 A0A182L730 A0A182XG44 Q7QB14 A0A182ID90 W8BY94 A0A182JPK1 A0A182SBM9 A0A0T6B8U0 A0A1B6G8J9 A0A182TLV2 V5RFY2 A0A084VBH0 A0A0L0BY42 A0A1I8Q4Q0 A0A182W1A2 A0A034WEB5 Q4AE76 A0A0K8WH30 A0A0A1WVM5 V5IAP2 Q298V2 B4G4Q0 A0A3B0KVS9 A0A2M3Z592 T1PFQ5 A0A232END1 K7J033 A0A182FPT9 T1DFU8 U5EWB8 A0A2M4BIZ6 A0A2M4A846 W5JPB5 A0A1W4WP68 B3LZZ2 J3JW40 A0A2A3E1J8 A0A088AVZ9 B4NJT3 B4MB83 B3P8P1 A0A1W4W414 A0A1L8DUD6 B4R030 A4V391 B4HEA1 P12613 B4PMA7 B4KC16 B4JSB6 A0A1A9WGN5 A0A1S3DVC9 A0A0T6AZW5 A0A1B0B2L3 A0A1A9YSY7 A0A067R174 A0A0K8TS69 A0A182G585 A0A336M5Y4 A0A158NJ24 A0A026WPB8 S4PEJ7 A0A1A9ZZ30 A0A212F181 D3TLP9 A0A1A9VFV7 E0VDT3 A0A182Y5D7 A0A1B6HYR1 A0A2J7QHG1 E9GKZ7 A0A0P5W1F5 A0A0P4ZAW1 A0A0P5TY06 A0A0M4ERP9 A0A0P5TRD2 A0A0P6FRS9
PDB
4B2T
E-value=0,
Score=1971
Ontologies
KEGG
GO
Topology
Subcellular location
Cytoplasm
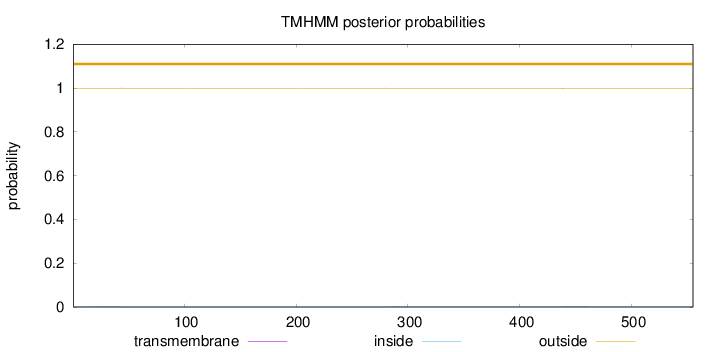
Length:
555
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.073
Exp number, first 60 AAs:
0.01102
Total prob of N-in:
0.00106
outside
1 - 555
Population Genetic Test Statistics
Pi
18.537183
Theta
16.015021
Tajima's D
0.585042
CLR
0.165525
CSRT
0.553372331383431
Interpretation
Uncertain
