Gene
KWMTBOMO03624
Pre Gene Modal
BGIBMGA006330
Annotation
PREDICTED:_serine/threonine-protein_kinase_Warts_[Papilio_xuthus]
Location in the cell
Nuclear Reliability : 3.964
Sequence
CDS
ATGGTGCGGAGGATAATAAACAAGGATATGTACAGGGTGTGGGGTTCATCAGCTGCTAGCACAGTTAGTTCAGGAGTGAGTTCGGGGTTTAGTTCTTCTTCGGGAAATGGACTCGATAAAGACTTGAATGTGCTACCTCAATCCTTGAACCAACTTATTGCCTTGGGTTATGATGAGGACCCAGCTGTGAGGGCACTCAAATATGCAGGAGGGCGTTTTGATGCAGCTCTCGATTATTTGTCGAAACAGCAAGAACCTCTCAACGGCGTCCTCAAAAGCAGTAACTTGAGTGCTCTTGGCACCAAACTTATCCGGAAACCTAGTCTAGAGAGAGAGATAAATCTACATCGTGGTAGTCCTGCACTTGATTCAGGTGCTGGTAGCTCCCGGTCTGATAGTCCGCGTCAATCGGAGCCACCGCCGCTACCACACGAGAAGCTATCGCGACAGTATTCGCCTTCAGGTTTCTCAGAACCACCGCCACCTCCGCCGCCGCGATGTCCGTCCACGCCGCCGGTGCTGCCTTCCGTGCAGCAACTGCTCAAGCGCATGTCGCCGGCGCCACCTCTACCACCGGCGCGGGGCACCAGTCCCGTGGCGGCCGCGGCACCCACGCCGCCTGCCAGACAACCCATGATTGTACAAAATGGACCTCAAGTGCAACAGCAGCTTACGCAGCAGATACAAGCTCTCAGTATCTATCAAACGGGTGGCGGCGGCGAACTTCCACCGCCGTATCCGATGTCGGGTGCGCCGCCACCTCCTCCCTACTCGGTCTCTATCCAAAATCGTCAAAGCCCAACCCAGTCGCAGGATTACAGGAAGAGTCCATCTTCGGGGATCTATTCAGGAGGCACCTCCGCCGGCTCGCCGAGTCCAATTACGGTAACGCAATCGACTGGTTCAGCCGCGGGAATGACTCGCCCCACGCCGCTCCAGGCGTGGACCGCGAGACAAGCGGTGCAGCCTCCTATCATCATGCAGTCTGTGAAAAGCACACAAGTTCAGAAACCGGTACTACAAACGGCGATAGCACCGGTTGCCCCACCGCCAGCCACCAGTGCCGGAGCACCCCCACCCCCTCCTTCGTACGCGAGCTCCATCCAACAGAAGCAGGCACAGACTCCGCCCAGCTACCCGTCGGCACCGAAGCCGTCGTCTCCTGGCTCCACACCTACTGCGATTCCTCCCGCCGTCCCGACCACGGAGCCACCAAGTTATGCGATCACGATGCAAGTGCTCGCCGTTCAACGCGGCATGCACCCCGTCCCGCCGCCTCCGTACGGTAACCAAGCCGACAACACGACTACTAACTGTACGAATAATCTCTTAAAAGACAACGTCACGGCCTCCGGCTCCGACAAAGGCTCTAACGGATCATCGGACAGACGACCAAAAATGTTAGATAAGATAAGGCATCAATCGCCAATACCGGAACGCAGAAATTACAGTAAAGAAAAAGAAGACGAGAGAAGAGATTGTAAAGTCCGAAATTATTCACCGCAAGCTTTCAAATTTTTTATGGAACAGCATGTGGAAAACGTTTTGAAATCTTATAAACAGAGACTGTTCAGAAGGATGCAACTCGAAAAGGAGATGAGTAAAATAGGTCTAAGTGCAGAAGCTCAGTGTCAGATGAGAAAGATGCTGTCCCAGAAGGAATCCAATTATATTCGATTGAAAAGGGCCAAGATGGACAAGTCTATGTTTACTAAGATTAAGCCTATAGGTGTGGGAGCATTTGGGGAGGTGACGTTGGTGAAAAAGATCGATACTAGTCATCTGTATGCCATGAAGACACTTCGGAAAGCTGACGTGCTAAAGAGAAATCAAGTGGCACACGTAAAAGCCGAAAGAGACATATTGGCGGAGGCAGACAACGAATGGGTCGTCAAGCTTTATTACAGTTTCCAAGATAAAGATAATCTGTATTTTGTAATGGACTATATACCGGGCGGGGACCTTATGTCCTTATTAATAAAATTAGGAATATTCGAGGAGAATCTCGCTAGGTTCTACATTGCAGAGTTGACGTGTGCTGTCGAAAGCGTGCACAAAATGGGTTTTATCCACAGAGACATCAAGCCAGATAACATATTGATAGATCGTGATGGTCATATAAAGTTAACAGATTTTGGTCTATGCACCGGTTTTAGATGGACGCACAACTCCAAATATTATCAAAGGAATGATCATGGACGACAAGACTCAATGGATCCAGTGGATGGAGAATGGGGGGCTATGGGCGAATGTCGATGTTATCAACTCAAACCCTTAGAAAGGCGGCGGAGAAGAGAACACCAGCGGTGTTTGGCCCATTCGTTAGTGGGAACCCCAAACTATATTGCGCCTGAAGTCTTGCAGCGGACTGGATACACGCAGCTTTGCGATTGGTGGTCGGTTGGAGTTATATTGTACGAGATGTTGGTTGGGTCGCCGCCGTTCCTGGCGCCGACCCCCGCAGAGACGCAACTCAAGGTGATCAATTGGGAGAGTACACTACACGTACCTGATGCAGCCAACTTATCCCCAGAGAGCAAAGACCTCATTCTGCAGCTATGTTCTGGTCAAGACACAAGGCTTGGCAAAGATGCCAATGAAGTAAAAAACCATCCCTTCCTCAAGGGCATCGATTTTGACAAAGGATTGAGGAATCAGGTTGCTCCTTACATCCCGAGGATTGAATACCCTACAGACACATCCAACTTCGACCCCATTGATCCAGATAAGCTCAGGAACTCTGGTAGTTCAGATTCAAATAAATCAGACAGTGAACTATTAGACAATGGCAAGACTTTCCATGGTTTCTTTGAGTTCACATTCAGAAGATTCTTTGATGATGGCTATACGAGCAAAATCAATTTGGATGACAATGACAATCAAGGACCTGTGTACGTGTGA
Protein
MVRRIINKDMYRVWGSSAASTVSSGVSSGFSSSSGNGLDKDLNVLPQSLNQLIALGYDEDPAVRALKYAGGRFDAALDYLSKQQEPLNGVLKSSNLSALGTKLIRKPSLEREINLHRGSPALDSGAGSSRSDSPRQSEPPPLPHEKLSRQYSPSGFSEPPPPPPPRCPSTPPVLPSVQQLLKRMSPAPPLPPARGTSPVAAAAPTPPARQPMIVQNGPQVQQQLTQQIQALSIYQTGGGGELPPPYPMSGAPPPPPYSVSIQNRQSPTQSQDYRKSPSSGIYSGGTSAGSPSPITVTQSTGSAAGMTRPTPLQAWTARQAVQPPIIMQSVKSTQVQKPVLQTAIAPVAPPPATSAGAPPPPPSYASSIQQKQAQTPPSYPSAPKPSSPGSTPTAIPPAVPTTEPPSYAITMQVLAVQRGMHPVPPPPYGNQADNTTTNCTNNLLKDNVTASGSDKGSNGSSDRRPKMLDKIRHQSPIPERRNYSKEKEDERRDCKVRNYSPQAFKFFMEQHVENVLKSYKQRLFRRMQLEKEMSKIGLSAEAQCQMRKMLSQKESNYIRLKRAKMDKSMFTKIKPIGVGAFGEVTLVKKIDTSHLYAMKTLRKADVLKRNQVAHVKAERDILAEADNEWVVKLYYSFQDKDNLYFVMDYIPGGDLMSLLIKLGIFEENLARFYIAELTCAVESVHKMGFIHRDIKPDNILIDRDGHIKLTDFGLCTGFRWTHNSKYYQRNDHGRQDSMDPVDGEWGAMGECRCYQLKPLERRRRREHQRCLAHSLVGTPNYIAPEVLQRTGYTQLCDWWSVGVILYEMLVGSPPFLAPTPAETQLKVINWESTLHVPDAANLSPESKDLILQLCSGQDTRLGKDANEVKNHPFLKGIDFDKGLRNQVAPYIPRIEYPTDTSNFDPIDPDKLRNSGSSDSNKSDSELLDNGKTFHGFFEFTFRRFFDDGYTSKINLDDNDNQGPVYV
Summary
Uniprot
EMBL
BABH01026063
NWSH01003141
PCG66875.1
KZ149893
PZC78927.1
KQ459011
+ More
KPJ04339.1 KQ460647 KPJ13172.1 PCG66874.1 KK855054 PTY20564.1 GEDC01023905 JAS13393.1 GEDC01005095 JAS32203.1 GEBQ01024796 GEBQ01007893 GEBQ01006404 GEBQ01001724 JAT15181.1 JAT32084.1 JAT33573.1 JAT38253.1 GEBQ01020742 GEBQ01015387 GEBQ01010212 GEBQ01002996 JAT19235.1 JAT24590.1 JAT29765.1 JAT36981.1 GECZ01017423 JAS52346.1 AJWK01022163 AJWK01022164 AJWK01022165 AJWK01022166 AJWK01022167
KPJ04339.1 KQ460647 KPJ13172.1 PCG66874.1 KK855054 PTY20564.1 GEDC01023905 JAS13393.1 GEDC01005095 JAS32203.1 GEBQ01024796 GEBQ01007893 GEBQ01006404 GEBQ01001724 JAT15181.1 JAT32084.1 JAT33573.1 JAT38253.1 GEBQ01020742 GEBQ01015387 GEBQ01010212 GEBQ01002996 JAT19235.1 JAT24590.1 JAT29765.1 JAT36981.1 GECZ01017423 JAS52346.1 AJWK01022163 AJWK01022164 AJWK01022165 AJWK01022166 AJWK01022167
Proteomes
PRIDE
Interpro
ProteinModelPortal
Ontologies
PATHWAY
GO
PANTHER
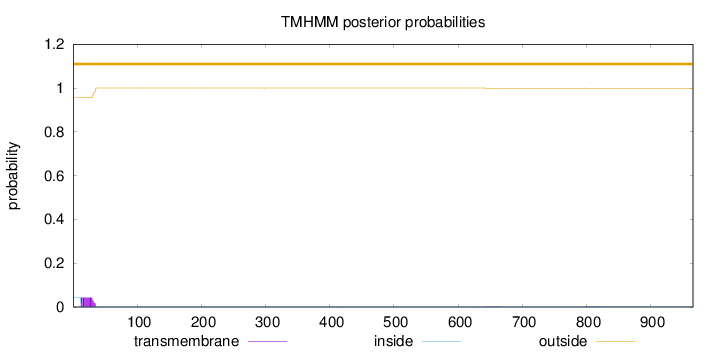
Topology
Length:
966
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.905569999999997
Exp number, first 60 AAs:
0.88358
Total prob of N-in:
0.04267
outside
1 - 966
Population Genetic Test Statistics
Pi
20.295591
Theta
17.036165
Tajima's D
0.07763
CLR
0.775613
CSRT
0.393180340982951
Interpretation
Uncertain
