Pre Gene Modal
BGIBMGA006265
Annotation
PREDICTED:_thioredoxin_domain-containing_protein_5_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.554 Extracellular Reliability : 1.091 PlasmaMembrane Reliability : 1.123
Sequence
CDS
ATGAAGATATTACCATATCTATTTGTATTTCCGTATCTTGTAGCTCCTGAACAGAGTTCTGTTTACGAATACAATCCCAGTAATTTCAAATTCCAGATCGAAGAAATGGATGGAAACTTTATTATGTTTTACGCGCCATGGTGTAGACATTGCACAGAATTTTATCCGATATGGTCAGAATTGGCAGAGCTGGTTAATACCAAGGACTCCAAATTTGCCATTGCCCAAGTTGACTGCACAGTGCATGCTAAGCTTTGTCATGAAAATGAAATAACAGGTTATCCAACATTATTTTATTTCCACAAAAATACATTTACACCAGTTGAATACAAAGGAACAAGGGACTTACCATCATTGACATTGTTTCTGAGCGAAGCCTTCTCTGTCAAAACAGAGGGCAAACAATCAAAACAACCAAATGAAGTTAAAACATACAGCGGCATGTCCTATCTGAATGACCTGAATATTGAGAAGTTTGTGTCAAAAGGACAGCATTTCATCATGTTTTTTGTACCTTGGTGTCGTGCATCTCAGAGGATGGCACCGATTTGGGCAGACTTAGCAGTACATTACGCTCACAACAACTACATAAAAATTGGAAAAGTTAATTGTATGGATAATGAGATAACATGCGAAAATTTTGAAGTCAAACAGTACCCCTATTTACTCTGGATTGTTAATGGAAAAATTATGGGTGCTTCGAATGGGGAAAACTTAGACGATTTGAAAGCATTTGTTGAAAAAATGTTGCTTTCTGAAAATCACGATCCAGAGAAATTTCTAAGAAAAAAGAAAGCACTTCCTGTGGCCAGGATATCTGAGGAGACATTCGAAACTTTTCTGCAAAATGACTTAGTTTTTATTAATTATTTTGCGCCATGGTGCGCACATTGTATGCAGTTAAGCCCATTATGGATTAAATTAGGGGAAAAGTTCCAAAACGAGACAAGAGTATTGATAGCAGATGTTGACTGTGTGCAGAGTAAACCTATTTGTGAAACAGAAAAGATTAATGGACTACCCACATTAATATTGTACAGACATAAGAAAATAGTAAGTGTAGAACATGGTGGTAGACCTCTTGATAGTCTGATTGGACTCATCACAGAGCATTTACAAGATTCAGCCAGTACAGATCCAGAAGACAAGACAAATGAGGACATATTTTTGTCAAAAGCCAAAGACGAATTGTAA
Protein
MKILPYLFVFPYLVAPEQSSVYEYNPSNFKFQIEEMDGNFIMFYAPWCRHCTEFYPIWSELAELVNTKDSKFAIAQVDCTVHAKLCHENEITGYPTLFYFHKNTFTPVEYKGTRDLPSLTLFLSEAFSVKTEGKQSKQPNEVKTYSGMSYLNDLNIEKFVSKGQHFIMFFVPWCRASQRMAPIWADLAVHYAHNNYIKIGKVNCMDNEITCENFEVKQYPYLLWIVNGKIMGASNGENLDDLKAFVEKMLLSENHDPEKFLRKKKALPVARISEETFETFLQNDLVFINYFAPWCAHCMQLSPLWIKLGEKFQNETRVLIADVDCVQSKPICETEKINGLPTLILYRHKKIVSVEHGGRPLDSLIGLITEHLQDSASTDPEDKTNEDIFLSKAKDEL
Summary
Similarity
Belongs to the protein disulfide isomerase family.
Uniprot
H9J9S2
A0A2A4J559
A0A2W1BV46
A0A194QFL9
A0A194R5S9
A0A0L7LQY7
+ More
A0A212ER45 A0A224XI07 V5GI69 T1HXQ3 A0A0K8SP98 A0A0A9XS29 A0A1W4WW63 D6WD23 A0A088AID6 E2B7N7 A0A0C9RKX2 A0A1B0CZI9 A0A2R7W1M4 A0A195FWA3 A0A1Y1K2X5 A0A151WV88 F4WNH5 F5B4Q2 A0A0P4WIM3 A0A293MWJ0 B7PHA0 A0A1S3DB21 E9FY51 A0A1L8DU05 A0A220XIQ9 U5ER38 A0A0P4ZYN5 A0A158NXZ4 A0A1B6DFB0 A0A0P5BIV9 A0A067QSV0 A0A0P5BPG9 A0A0P5WTL7 A0A0P5ZTP5 A0A0P5KFW1 A0A0P6CVG8 A0A0P6E4V3 A0A0P5ZLB6 A0A0P5TR08 A0A0P5CTX2 A0A0P5XH58 A0A0P5BPG7 A0A0T6B8U2 A0A1Q3F9P0 A0A2A3EJY6 A0A0P5B5Q6 A0A0P4WMZ7 Q17I04 A0A1S4F1W0 A0A164M4U4 A0A0P5SYM4 A0A0P5FJD1 A0A023ESN4 A0A2J7QRB2 A0A0P5DBJ1 A0A2R5LFZ4 A0A1L8EID4 B3MQD1 A0A0L0BRP8 A0A1I8PHM3 Q1HR90 U4UU64 A0A1L8EEK6 N6TSU7 A0A1A9VRC5 A0A0P5ZUX1 A0A0P5CYL0 A0A1A9XDU9 A0A1B0B8G6 A0A023F8J0 R4G5U7 A0A023EQD7 A0A2T7PG55 A0A0L7QVY2 A0A1B0A064 A0A0M4EUS3 J3JWJ1 E0VDX9 A0A0P5VHN2 A0A0P5VI97 V4AG03 A0A195ECZ0 A0A1W4VZU0 B3NU99 A0A1A9WCI7 A0A3B0K196 B4NDP8 Q29JA8 A0A151IM81 A0A1I8NGE4 B4R365 A0A1J1ICW5 A0A1J1I965
A0A212ER45 A0A224XI07 V5GI69 T1HXQ3 A0A0K8SP98 A0A0A9XS29 A0A1W4WW63 D6WD23 A0A088AID6 E2B7N7 A0A0C9RKX2 A0A1B0CZI9 A0A2R7W1M4 A0A195FWA3 A0A1Y1K2X5 A0A151WV88 F4WNH5 F5B4Q2 A0A0P4WIM3 A0A293MWJ0 B7PHA0 A0A1S3DB21 E9FY51 A0A1L8DU05 A0A220XIQ9 U5ER38 A0A0P4ZYN5 A0A158NXZ4 A0A1B6DFB0 A0A0P5BIV9 A0A067QSV0 A0A0P5BPG9 A0A0P5WTL7 A0A0P5ZTP5 A0A0P5KFW1 A0A0P6CVG8 A0A0P6E4V3 A0A0P5ZLB6 A0A0P5TR08 A0A0P5CTX2 A0A0P5XH58 A0A0P5BPG7 A0A0T6B8U2 A0A1Q3F9P0 A0A2A3EJY6 A0A0P5B5Q6 A0A0P4WMZ7 Q17I04 A0A1S4F1W0 A0A164M4U4 A0A0P5SYM4 A0A0P5FJD1 A0A023ESN4 A0A2J7QRB2 A0A0P5DBJ1 A0A2R5LFZ4 A0A1L8EID4 B3MQD1 A0A0L0BRP8 A0A1I8PHM3 Q1HR90 U4UU64 A0A1L8EEK6 N6TSU7 A0A1A9VRC5 A0A0P5ZUX1 A0A0P5CYL0 A0A1A9XDU9 A0A1B0B8G6 A0A023F8J0 R4G5U7 A0A023EQD7 A0A2T7PG55 A0A0L7QVY2 A0A1B0A064 A0A0M4EUS3 J3JWJ1 E0VDX9 A0A0P5VHN2 A0A0P5VI97 V4AG03 A0A195ECZ0 A0A1W4VZU0 B3NU99 A0A1A9WCI7 A0A3B0K196 B4NDP8 Q29JA8 A0A151IM81 A0A1I8NGE4 B4R365 A0A1J1ICW5 A0A1J1I965
Pubmed
EMBL
BABH01026061
BABH01026062
BABH01026063
NWSH01003141
PCG66876.1
KZ149893
+ More
PZC78928.1 KQ459011 KPJ04338.1 KQ460647 KPJ13173.1 JTDY01000294 KOB77845.1 AGBW02013152 OWR43931.1 GFTR01005742 JAW10684.1 GALX01004717 JAB63749.1 ACPB03021026 GBRD01010781 JAG55043.1 GBHO01020850 JAG22754.1 KQ971311 EEZ99095.1 GL446193 EFN88315.1 GBYB01008830 JAG78597.1 AJVK01020820 KK854227 PTY13338.1 KQ981215 KYN44597.1 GEZM01098817 JAV53736.1 KQ982706 KYQ51850.1 GL888237 EGI64168.1 HQ630061 AEE36485.1 GDRN01070603 GDRN01070602 JAI63841.1 GFWV01018787 MAA43515.1 ABJB010253034 ABJB010429163 ABJB010654407 ABJB010897971 ABJB011140338 DS711754 EEC05972.1 GL732527 EFX87477.1 GFDF01004152 JAV09932.1 MF278684 ASL04990.1 GANO01002981 JAB56890.1 GDIP01206344 JAJ17058.1 ADTU01002818 GEDC01012917 JAS24381.1 GDIP01189243 JAJ34159.1 KK853582 KDR06511.1 GDIP01181741 JAJ41661.1 GDIP01082653 JAM21062.1 GDIP01051119 GDIP01039964 JAM63751.1 GDIQ01210412 JAK41313.1 GDIQ01086808 JAN07929.1 GDIP01252576 GDIQ01068923 JAI70825.1 JAN25814.1 GDIP01147750 GDIP01045722 JAM57993.1 GDIP01158899 GDIP01140754 GDIQ01068922 JAL62960.1 JAN25815.1 GDIP01166589 JAJ56813.1 GDIP01084507 JAM19208.1 GDIP01181740 JAJ41662.1 LJIG01009148 KRT83659.1 GFDL01010741 JAV24304.1 KZ288222 PBC32037.1 GDIP01189246 JAJ34156.1 GDIP01254580 JAI68821.1 CH477244 EAT46272.1 LRGB01003089 KZS04728.1 GDIP01133601 JAL70113.1 GDIQ01253760 JAJ97964.1 GAPW01001802 JAC11796.1 NEVH01011914 PNF31129.1 GDIP01163749 JAJ59653.1 GGLE01004308 MBY08434.1 GFDG01000385 JAV18414.1 CH902621 EDV44557.1 JRES01001467 KNC22673.1 DQ440204 ABF18237.1 KI209717 KI210120 KI210134 ERL96078.1 ERL96160.1 ERL96167.1 GFDG01001678 JAV17121.1 APGK01053332 KB741223 ENN72350.1 GDIP01038713 JAM65002.1 GDIP01163748 JAJ59654.1 JXJN01009934 GBBI01001052 JAC17660.1 GAHY01000210 JAA77300.1 GAPW01001801 JAC11797.1 PZQS01000004 PVD32417.1 KQ414721 KOC62772.1 CP012528 ALC48885.1 BT127609 AEE62571.1 DS235088 EEB11585.1 GDIP01099800 JAM03915.1 GDIP01099801 JAM03914.1 KB201864 ESO94095.1 KQ979074 KYN22709.1 CH954180 EDV46014.1 OUUW01000003 SPP78791.1 CH964239 EDW81870.1 CH379063 EAL32393.1 KQ977063 KYN06001.1 CM000366 EDX17667.1 CVRI01000044 CRK96814.1 CRK96813.1
PZC78928.1 KQ459011 KPJ04338.1 KQ460647 KPJ13173.1 JTDY01000294 KOB77845.1 AGBW02013152 OWR43931.1 GFTR01005742 JAW10684.1 GALX01004717 JAB63749.1 ACPB03021026 GBRD01010781 JAG55043.1 GBHO01020850 JAG22754.1 KQ971311 EEZ99095.1 GL446193 EFN88315.1 GBYB01008830 JAG78597.1 AJVK01020820 KK854227 PTY13338.1 KQ981215 KYN44597.1 GEZM01098817 JAV53736.1 KQ982706 KYQ51850.1 GL888237 EGI64168.1 HQ630061 AEE36485.1 GDRN01070603 GDRN01070602 JAI63841.1 GFWV01018787 MAA43515.1 ABJB010253034 ABJB010429163 ABJB010654407 ABJB010897971 ABJB011140338 DS711754 EEC05972.1 GL732527 EFX87477.1 GFDF01004152 JAV09932.1 MF278684 ASL04990.1 GANO01002981 JAB56890.1 GDIP01206344 JAJ17058.1 ADTU01002818 GEDC01012917 JAS24381.1 GDIP01189243 JAJ34159.1 KK853582 KDR06511.1 GDIP01181741 JAJ41661.1 GDIP01082653 JAM21062.1 GDIP01051119 GDIP01039964 JAM63751.1 GDIQ01210412 JAK41313.1 GDIQ01086808 JAN07929.1 GDIP01252576 GDIQ01068923 JAI70825.1 JAN25814.1 GDIP01147750 GDIP01045722 JAM57993.1 GDIP01158899 GDIP01140754 GDIQ01068922 JAL62960.1 JAN25815.1 GDIP01166589 JAJ56813.1 GDIP01084507 JAM19208.1 GDIP01181740 JAJ41662.1 LJIG01009148 KRT83659.1 GFDL01010741 JAV24304.1 KZ288222 PBC32037.1 GDIP01189246 JAJ34156.1 GDIP01254580 JAI68821.1 CH477244 EAT46272.1 LRGB01003089 KZS04728.1 GDIP01133601 JAL70113.1 GDIQ01253760 JAJ97964.1 GAPW01001802 JAC11796.1 NEVH01011914 PNF31129.1 GDIP01163749 JAJ59653.1 GGLE01004308 MBY08434.1 GFDG01000385 JAV18414.1 CH902621 EDV44557.1 JRES01001467 KNC22673.1 DQ440204 ABF18237.1 KI209717 KI210120 KI210134 ERL96078.1 ERL96160.1 ERL96167.1 GFDG01001678 JAV17121.1 APGK01053332 KB741223 ENN72350.1 GDIP01038713 JAM65002.1 GDIP01163748 JAJ59654.1 JXJN01009934 GBBI01001052 JAC17660.1 GAHY01000210 JAA77300.1 GAPW01001801 JAC11797.1 PZQS01000004 PVD32417.1 KQ414721 KOC62772.1 CP012528 ALC48885.1 BT127609 AEE62571.1 DS235088 EEB11585.1 GDIP01099800 JAM03915.1 GDIP01099801 JAM03914.1 KB201864 ESO94095.1 KQ979074 KYN22709.1 CH954180 EDV46014.1 OUUW01000003 SPP78791.1 CH964239 EDW81870.1 CH379063 EAL32393.1 KQ977063 KYN06001.1 CM000366 EDX17667.1 CVRI01000044 CRK96814.1 CRK96813.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000037510
UP000007151
+ More
UP000015103 UP000192223 UP000007266 UP000005203 UP000008237 UP000092462 UP000078541 UP000075809 UP000007755 UP000001555 UP000079169 UP000000305 UP000005205 UP000027135 UP000242457 UP000008820 UP000076858 UP000235965 UP000007801 UP000037069 UP000095300 UP000030742 UP000019118 UP000078200 UP000092443 UP000092460 UP000245119 UP000053825 UP000092445 UP000092553 UP000009046 UP000030746 UP000078492 UP000192221 UP000008711 UP000091820 UP000268350 UP000007798 UP000001819 UP000078542 UP000095301 UP000000304 UP000183832
UP000015103 UP000192223 UP000007266 UP000005203 UP000008237 UP000092462 UP000078541 UP000075809 UP000007755 UP000001555 UP000079169 UP000000305 UP000005205 UP000027135 UP000242457 UP000008820 UP000076858 UP000235965 UP000007801 UP000037069 UP000095300 UP000030742 UP000019118 UP000078200 UP000092443 UP000092460 UP000245119 UP000053825 UP000092445 UP000092553 UP000009046 UP000030746 UP000078492 UP000192221 UP000008711 UP000091820 UP000268350 UP000007798 UP000001819 UP000078542 UP000095301 UP000000304 UP000183832
PRIDE
Interpro
IPR036249
Thioredoxin-like_sf
+ More
IPR013766 Thioredoxin_domain
IPR017937 Thioredoxin_CS
IPR005788 Disulphide_isomerase
IPR020103 PsdUridine_synth_cat_dom_sf
IPR016193 Cytidine_deaminase-like
IPR002125 CMP_dCMP_dom
IPR029021 Prot-tyrosine_phosphatase-like
IPR016130 Tyr_Pase_AS
IPR000387 TYR_PHOSPHATASE_dom
IPR000340 Dual-sp_phosphatase_cat-dom
IPR014876 DEK_C
IPR020422 TYR_PHOSPHATASE_DUAL_dom
IPR013766 Thioredoxin_domain
IPR017937 Thioredoxin_CS
IPR005788 Disulphide_isomerase
IPR020103 PsdUridine_synth_cat_dom_sf
IPR016193 Cytidine_deaminase-like
IPR002125 CMP_dCMP_dom
IPR029021 Prot-tyrosine_phosphatase-like
IPR016130 Tyr_Pase_AS
IPR000387 TYR_PHOSPHATASE_dom
IPR000340 Dual-sp_phosphatase_cat-dom
IPR014876 DEK_C
IPR020422 TYR_PHOSPHATASE_DUAL_dom
Gene 3D
CDD
ProteinModelPortal
H9J9S2
A0A2A4J559
A0A2W1BV46
A0A194QFL9
A0A194R5S9
A0A0L7LQY7
+ More
A0A212ER45 A0A224XI07 V5GI69 T1HXQ3 A0A0K8SP98 A0A0A9XS29 A0A1W4WW63 D6WD23 A0A088AID6 E2B7N7 A0A0C9RKX2 A0A1B0CZI9 A0A2R7W1M4 A0A195FWA3 A0A1Y1K2X5 A0A151WV88 F4WNH5 F5B4Q2 A0A0P4WIM3 A0A293MWJ0 B7PHA0 A0A1S3DB21 E9FY51 A0A1L8DU05 A0A220XIQ9 U5ER38 A0A0P4ZYN5 A0A158NXZ4 A0A1B6DFB0 A0A0P5BIV9 A0A067QSV0 A0A0P5BPG9 A0A0P5WTL7 A0A0P5ZTP5 A0A0P5KFW1 A0A0P6CVG8 A0A0P6E4V3 A0A0P5ZLB6 A0A0P5TR08 A0A0P5CTX2 A0A0P5XH58 A0A0P5BPG7 A0A0T6B8U2 A0A1Q3F9P0 A0A2A3EJY6 A0A0P5B5Q6 A0A0P4WMZ7 Q17I04 A0A1S4F1W0 A0A164M4U4 A0A0P5SYM4 A0A0P5FJD1 A0A023ESN4 A0A2J7QRB2 A0A0P5DBJ1 A0A2R5LFZ4 A0A1L8EID4 B3MQD1 A0A0L0BRP8 A0A1I8PHM3 Q1HR90 U4UU64 A0A1L8EEK6 N6TSU7 A0A1A9VRC5 A0A0P5ZUX1 A0A0P5CYL0 A0A1A9XDU9 A0A1B0B8G6 A0A023F8J0 R4G5U7 A0A023EQD7 A0A2T7PG55 A0A0L7QVY2 A0A1B0A064 A0A0M4EUS3 J3JWJ1 E0VDX9 A0A0P5VHN2 A0A0P5VI97 V4AG03 A0A195ECZ0 A0A1W4VZU0 B3NU99 A0A1A9WCI7 A0A3B0K196 B4NDP8 Q29JA8 A0A151IM81 A0A1I8NGE4 B4R365 A0A1J1ICW5 A0A1J1I965
A0A212ER45 A0A224XI07 V5GI69 T1HXQ3 A0A0K8SP98 A0A0A9XS29 A0A1W4WW63 D6WD23 A0A088AID6 E2B7N7 A0A0C9RKX2 A0A1B0CZI9 A0A2R7W1M4 A0A195FWA3 A0A1Y1K2X5 A0A151WV88 F4WNH5 F5B4Q2 A0A0P4WIM3 A0A293MWJ0 B7PHA0 A0A1S3DB21 E9FY51 A0A1L8DU05 A0A220XIQ9 U5ER38 A0A0P4ZYN5 A0A158NXZ4 A0A1B6DFB0 A0A0P5BIV9 A0A067QSV0 A0A0P5BPG9 A0A0P5WTL7 A0A0P5ZTP5 A0A0P5KFW1 A0A0P6CVG8 A0A0P6E4V3 A0A0P5ZLB6 A0A0P5TR08 A0A0P5CTX2 A0A0P5XH58 A0A0P5BPG7 A0A0T6B8U2 A0A1Q3F9P0 A0A2A3EJY6 A0A0P5B5Q6 A0A0P4WMZ7 Q17I04 A0A1S4F1W0 A0A164M4U4 A0A0P5SYM4 A0A0P5FJD1 A0A023ESN4 A0A2J7QRB2 A0A0P5DBJ1 A0A2R5LFZ4 A0A1L8EID4 B3MQD1 A0A0L0BRP8 A0A1I8PHM3 Q1HR90 U4UU64 A0A1L8EEK6 N6TSU7 A0A1A9VRC5 A0A0P5ZUX1 A0A0P5CYL0 A0A1A9XDU9 A0A1B0B8G6 A0A023F8J0 R4G5U7 A0A023EQD7 A0A2T7PG55 A0A0L7QVY2 A0A1B0A064 A0A0M4EUS3 J3JWJ1 E0VDX9 A0A0P5VHN2 A0A0P5VI97 V4AG03 A0A195ECZ0 A0A1W4VZU0 B3NU99 A0A1A9WCI7 A0A3B0K196 B4NDP8 Q29JA8 A0A151IM81 A0A1I8NGE4 B4R365 A0A1J1ICW5 A0A1J1I965
PDB
2DIZ
E-value=1.57524e-17,
Score=219
Ontologies
GO
Topology
SignalP
Position: 1 - 16,
Likelihood: 0.864212
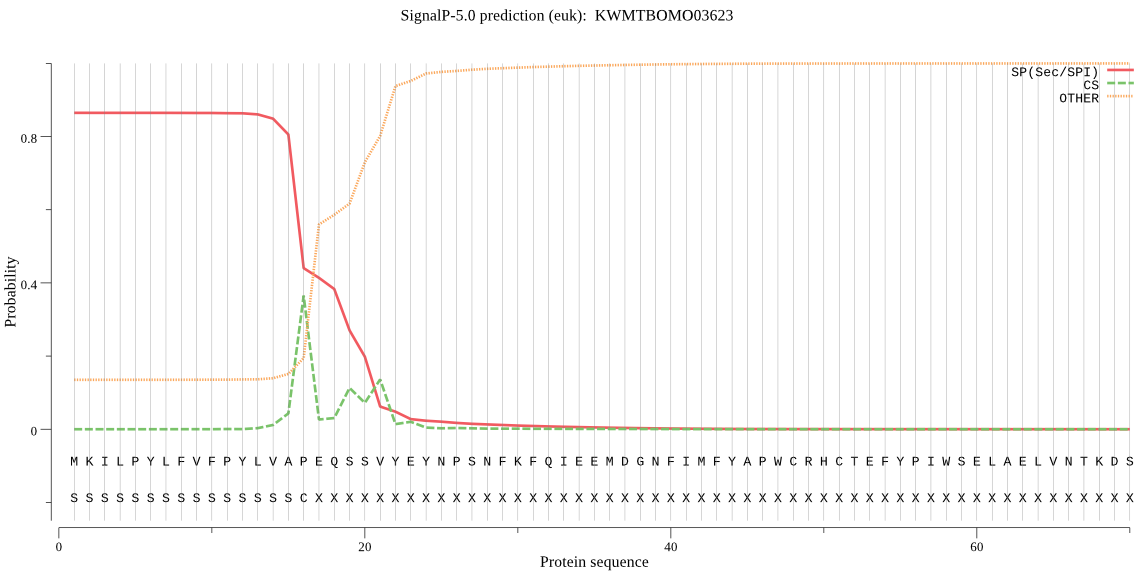
Length:
397
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.03082
Exp number, first 60 AAs:
0.00362
Total prob of N-in:
0.00117
outside
1 - 397
Population Genetic Test Statistics
Pi
18.373807
Theta
23.357917
Tajima's D
-0.521207
CLR
10.305241
CSRT
0.234888255587221
Interpretation
Uncertain
