Pre Gene Modal
BGIBMGA006327
Annotation
PREDICTED:_ankyrin_repeat_domain-containing_protein_17-like_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.717 Nuclear Reliability : 1.233
Sequence
CDS
ATGCAGAATGTTGGACAGTCTGAAAACCGAAATATCGATAAGGTGAATGTGCAACTTGATGTGGTAAAAACGTCTACTCCACAGAGTTCGGCTTCAAGTCCGGCCAAATCTGAAACTGAAACGTATTCAGGAGCTCCTGCCAAATATATGACAGACTCTTCGGAAAGTGAGGATGACTCCGTGTCTGAGGCCGAAATGGAAGAAGAAAATGGTAGTACCGAGCCATCTAAGTTCCTACTCTCGCACGATGATCCCGAAAGAGCTGTCGATCCTGAAATGCAAGCCAGACTGGAAGCATTATTAGAGGCAGCTGGTATTGGTAAACTGTCAGGCGAAGGCAAACACTTGGCCGACCCGGAGGTCCTTCGTAGGCTCACCTCTTCAGTTTCTTGCGCACTAGACGAGGCGGCCGCCGCCCTAACGAGAATGCGGAGCGACCAGCCCGCGCCCCACCACAGGCATCATCACCAAGAGAAGAATTTCAGTCAGCCGTGCGGGGTGACTGCGACGGGCTCGACGCCTGCGGTCCCCTCGTCTGTGGGCGCGGACGGAGCGCCGTCGCTTGTCGAGGCGTGCTCTGACGGCGACGTGGGCACTGTCCGCAAGCTACTCACCGAAGGGCGGTCTGTCCACGAGACGACCGAGGAAGGCGAGCCCTTACTCTCCCTCGCCTGCTCAGCGGGATATTACGAGCTCGCGCAGGTCCTCTTGGCGATGCACGCCAGCGTCGAGGACCGGGGTATCAAGGGCGACTGCACGCCGCTGATGGAGGCGGCGAGCGCCGGCCACGTGGACATCGTGCGGCTGCTGCTGGCGCACGGCGCGGACGTGAACGCGGTGTCCGGCTCCGGGAACACGCCGCTCATGTACGCGTGCGCGGGCGGACACGAGGACTGCGTGCGCGCGCTGCTCGACAACGGGGCTAACGTCGAGGACCACAACGAGAACGGGCACACGCCGCTCATGGAGGCCGCGTCGGCGGGCCATGTCGGCGTTGCAAAGATCCTGTTGGAGCACGGAGCAGGGATCAACACTCATTCTAACGAGTTCAAGGAATCCGCGTTAACGCTCGCCTGCTATAAGGGTCACCTGGACATGGTGCGGTTCCTGCTGGCGGCGGGCGCGGACCGCGAGCACAAGACGGACGAGATGCACACCGCGCTCATGGAGGCCAGCATGGACGGACACGTCGAGGTCGCCAGGCTGCTGCTCGACTCCGGGGCGCAGGTGAACATGCCAACGGATAGTTTCGAGTCGCCATTAACACTAGCGGCGTGTGGAGGCCACGTGGACCTGGCGATGTTGCTGCTGGAACGCGGCGCCAACATCGAGGAGGTCAATGACGAGGGCTACACGCCGCTCATGGAGGCCGCTAGGGAAGGTCACGAGGAGATGGTTGCGCTGCTGCTGGGTCAAGGTGCACAGATAAATGCACAGACCGAGGAAACGCAGGAGACTGCGCTGACCCTCGCCTGCTGCGGAGGGTTTCTCGAGGTGGCGGAATCGCTCATCAAGGCGGGCGCTGACGCTGAGCTCGGCGCTTCCACTCCGCTCATGGAGGCTTCGCAGGAAGGGCATCTAGAACTAGTACGGTATCTGATCAATGCGGGCGCGGACGTGCACGCGACGACGCAGACGGGCGACACGGCGCTGACGTACGCCTGCGAGAACGGGCACACGGACGTGGCGGAGCTGCTGCTGCGCGCCGGCGCCGCGCTGGAGCACGAGAGCGAGGGCGGCCGCACGCCCCTCATGAAGGCGTGTCGCGCCGGACACCTCTGCACCGTGCAGTTCCTAGTCGCGAAGGGGGCGGACGTTAACCGCATGACGGCTAGCGGCGACCACACGCCGTTGTCTCTGGCGTGCGCGGGCGGACACGTGGACGTGGTGAAGTTCTTGTTGGCCTGCGACGCCGATCCCTTCCGAAAACTCAAGGACAACTCCACCATGCTCATCGAGGCAGCCAAGGGCGGACACACGGCCGTCGTGCAACTGCTCCTCGACTTCCCACATTCTGTAATGCTTCCTAGAGGCACGACAACAACAACGACGGCGGAGGACGGCGCGGGGCTGAGCGCGGCGCAGGCGGCGGCGCTGGGGCTCACGCACGCGCCCGCCCCCGGCGCGCCCGCCGCCCGCGCGCTCCTGCCCGCGCACGCGCCCGCTCCCGACCTGCCACACAACTTTGCCAAGGCCTATCTCGATGAACGCAGTTCGGGCGGCGGTCGCAAGAAACCGAGCCCGTACCCAACGGCGGCGGCGGTGGCGAGCGCAATCTGCGGCGCGACGGGCAGGGCTGAGGCGTTGGTCGCGCAGCAGAGGAATGAATCGATGCCGGACACCGAGAGGGGGCTCTCGCTCGAAATTAACGATCCTTCAGGTCCCCCGACGTTGGCCACCTCGGCGCTGCACGCGCTCGACCTGCAGTTCTGCGCGCAAGGTGTCACAACGAGTCATTCGCCGGCCACTCCGCCGTACCCTCCTCCGCAACAATTGTTCCCAGTGCAACCAATACCGACCAACCTTAATCAGGTATTTTGA
Protein
MQNVGQSENRNIDKVNVQLDVVKTSTPQSSASSPAKSETETYSGAPAKYMTDSSESEDDSVSEAEMEEENGSTEPSKFLLSHDDPERAVDPEMQARLEALLEAAGIGKLSGEGKHLADPEVLRRLTSSVSCALDEAAAALTRMRSDQPAPHHRHHHQEKNFSQPCGVTATGSTPAVPSSVGADGAPSLVEACSDGDVGTVRKLLTEGRSVHETTEEGEPLLSLACSAGYYELAQVLLAMHASVEDRGIKGDCTPLMEAASAGHVDIVRLLLAHGADVNAVSGSGNTPLMYACAGGHEDCVRALLDNGANVEDHNENGHTPLMEAASAGHVGVAKILLEHGAGINTHSNEFKESALTLACYKGHLDMVRFLLAAGADREHKTDEMHTALMEASMDGHVEVARLLLDSGAQVNMPTDSFESPLTLAACGGHVDLAMLLLERGANIEEVNDEGYTPLMEAAREGHEEMVALLLGQGAQINAQTEETQETALTLACCGGFLEVAESLIKAGADAELGASTPLMEASQEGHLELVRYLINAGADVHATTQTGDTALTYACENGHTDVAELLLRAGAALEHESEGGRTPLMKACRAGHLCTVQFLVAKGADVNRMTASGDHTPLSLACAGGHVDVVKFLLACDADPFRKLKDNSTMLIEAAKGGHTAVVQLLLDFPHSVMLPRGTTTTTTAEDGAGLSAAQAAALGLTHAPAPGAPAARALLPAHAPAPDLPHNFAKAYLDERSSGGGRKKPSPYPTAAAVASAICGATGRAEALVAQQRNESMPDTERGLSLEINDPSGPPTLATSALHALDLQFCAQGVTTSHSPATPPYPPPQQLFPVQPIPTNLNQVF
Summary
Uniprot
S4PCZ3
A0A2H1WU13
A0A2A4JTS2
A0A2A4JTK0
A0A194R5T8
A0A2J7PZL6
+ More
A0A151WTV0 A0A195F6K1 A0A158NYD8 A0A195AYV4 F4WSY2 E2A2P9 A0A151J537 A0A0J7KN53 E2B848 A0A2A3EE44 A0A0M8ZW86 A0A088AN05 A0A026WBW5 A0A3L8DVI7 A0A0C9QWA5 A0A0C9R5G4 A0A0C9RM13 A0A154PAU1 A0A0C9RM09 A0A0C9Q1Z2 A0A1B6FPS8 A0A1B6G072 A0A1B6GWU8 A0A1B6G0W3 A0A310SQB5 A0A0A9WC50 A0A0A9W4V5 A0A0A9W1Y3 A0A0A9W3N6 A0A1Y1N2D3 A0A0A9XWA2 A0A1Y1MZF8 T1I0B8 D6W9Q4 V5GJL5 V5G2P6 E0VW24 A0A0P4VWP1 T1IU24 A0A224XG59 A0A069DYE2 A0A0V0G3H5 J9K5L3 A0A2H8TY68 A0A210PF93 A0A2S2Q793 A0A2S2NPD4 A0A2R2MPW2
A0A151WTV0 A0A195F6K1 A0A158NYD8 A0A195AYV4 F4WSY2 E2A2P9 A0A151J537 A0A0J7KN53 E2B848 A0A2A3EE44 A0A0M8ZW86 A0A088AN05 A0A026WBW5 A0A3L8DVI7 A0A0C9QWA5 A0A0C9R5G4 A0A0C9RM13 A0A154PAU1 A0A0C9RM09 A0A0C9Q1Z2 A0A1B6FPS8 A0A1B6G072 A0A1B6GWU8 A0A1B6G0W3 A0A310SQB5 A0A0A9WC50 A0A0A9W4V5 A0A0A9W1Y3 A0A0A9W3N6 A0A1Y1N2D3 A0A0A9XWA2 A0A1Y1MZF8 T1I0B8 D6W9Q4 V5GJL5 V5G2P6 E0VW24 A0A0P4VWP1 T1IU24 A0A224XG59 A0A069DYE2 A0A0V0G3H5 J9K5L3 A0A2H8TY68 A0A210PF93 A0A2S2Q793 A0A2S2NPD4 A0A2R2MPW2
Pubmed
EMBL
GAIX01003871
JAA88689.1
ODYU01011004
SOQ56472.1
NWSH01000685
PCG74792.1
+ More
PCG74793.1 KQ460647 KPJ13183.1 NEVH01020334 PNF21762.1 KQ982750 KYQ51266.1 KQ981768 KYN36011.1 ADTU01003765 ADTU01003766 KQ976701 KYM77222.1 GL888331 EGI62686.1 GL436195 EFN72287.1 KQ980055 KYN17976.1 LBMM01005181 KMQ91742.1 GL446282 EFN88140.1 KZ288269 PBC30025.1 KQ435824 KOX72207.1 KK107295 EZA53111.1 QOIP01000003 RLU24322.1 GBYB01008019 JAG77786.1 GBYB01008017 JAG77784.1 GBYB01008021 JAG77788.1 KQ434864 KZC09056.1 GBYB01008016 JAG77783.1 GBYB01008018 JAG77785.1 GECZ01017746 JAS52023.1 GECZ01013922 JAS55847.1 GECZ01002882 JAS66887.1 GECZ01013728 JAS56041.1 KQ761768 OAD57115.1 GBHO01041164 JAG02440.1 GBHO01041163 JAG02441.1 GBHO01041162 JAG02442.1 GBHO01041165 JAG02439.1 GEZM01016909 JAV91050.1 GBHO01019410 JAG24194.1 GEZM01016908 JAV91051.1 ACPB03019371 ACPB03019372 KQ971312 EEZ98529.2 GALX01004177 JAB64289.1 GALX01004176 JAB64290.1 DS235816 EEB17580.1 GDKW01000062 JAI56533.1 JH431516 GFTR01008986 JAW07440.1 GBGD01000013 JAC88876.1 GECL01003487 JAP02637.1 ABLF02024338 GFXV01006667 MBW18472.1 NEDP02076739 OWF35158.1 GGMS01004422 MBY73625.1 GGMR01006193 MBY18812.1
PCG74793.1 KQ460647 KPJ13183.1 NEVH01020334 PNF21762.1 KQ982750 KYQ51266.1 KQ981768 KYN36011.1 ADTU01003765 ADTU01003766 KQ976701 KYM77222.1 GL888331 EGI62686.1 GL436195 EFN72287.1 KQ980055 KYN17976.1 LBMM01005181 KMQ91742.1 GL446282 EFN88140.1 KZ288269 PBC30025.1 KQ435824 KOX72207.1 KK107295 EZA53111.1 QOIP01000003 RLU24322.1 GBYB01008019 JAG77786.1 GBYB01008017 JAG77784.1 GBYB01008021 JAG77788.1 KQ434864 KZC09056.1 GBYB01008016 JAG77783.1 GBYB01008018 JAG77785.1 GECZ01017746 JAS52023.1 GECZ01013922 JAS55847.1 GECZ01002882 JAS66887.1 GECZ01013728 JAS56041.1 KQ761768 OAD57115.1 GBHO01041164 JAG02440.1 GBHO01041163 JAG02441.1 GBHO01041162 JAG02442.1 GBHO01041165 JAG02439.1 GEZM01016909 JAV91050.1 GBHO01019410 JAG24194.1 GEZM01016908 JAV91051.1 ACPB03019371 ACPB03019372 KQ971312 EEZ98529.2 GALX01004177 JAB64289.1 GALX01004176 JAB64290.1 DS235816 EEB17580.1 GDKW01000062 JAI56533.1 JH431516 GFTR01008986 JAW07440.1 GBGD01000013 JAC88876.1 GECL01003487 JAP02637.1 ABLF02024338 GFXV01006667 MBW18472.1 NEDP02076739 OWF35158.1 GGMS01004422 MBY73625.1 GGMR01006193 MBY18812.1
Proteomes
Interpro
Gene 3D
CDD
ProteinModelPortal
S4PCZ3
A0A2H1WU13
A0A2A4JTS2
A0A2A4JTK0
A0A194R5T8
A0A2J7PZL6
+ More
A0A151WTV0 A0A195F6K1 A0A158NYD8 A0A195AYV4 F4WSY2 E2A2P9 A0A151J537 A0A0J7KN53 E2B848 A0A2A3EE44 A0A0M8ZW86 A0A088AN05 A0A026WBW5 A0A3L8DVI7 A0A0C9QWA5 A0A0C9R5G4 A0A0C9RM13 A0A154PAU1 A0A0C9RM09 A0A0C9Q1Z2 A0A1B6FPS8 A0A1B6G072 A0A1B6GWU8 A0A1B6G0W3 A0A310SQB5 A0A0A9WC50 A0A0A9W4V5 A0A0A9W1Y3 A0A0A9W3N6 A0A1Y1N2D3 A0A0A9XWA2 A0A1Y1MZF8 T1I0B8 D6W9Q4 V5GJL5 V5G2P6 E0VW24 A0A0P4VWP1 T1IU24 A0A224XG59 A0A069DYE2 A0A0V0G3H5 J9K5L3 A0A2H8TY68 A0A210PF93 A0A2S2Q793 A0A2S2NPD4 A0A2R2MPW2
A0A151WTV0 A0A195F6K1 A0A158NYD8 A0A195AYV4 F4WSY2 E2A2P9 A0A151J537 A0A0J7KN53 E2B848 A0A2A3EE44 A0A0M8ZW86 A0A088AN05 A0A026WBW5 A0A3L8DVI7 A0A0C9QWA5 A0A0C9R5G4 A0A0C9RM13 A0A154PAU1 A0A0C9RM09 A0A0C9Q1Z2 A0A1B6FPS8 A0A1B6G072 A0A1B6GWU8 A0A1B6G0W3 A0A310SQB5 A0A0A9WC50 A0A0A9W4V5 A0A0A9W1Y3 A0A0A9W3N6 A0A1Y1N2D3 A0A0A9XWA2 A0A1Y1MZF8 T1I0B8 D6W9Q4 V5GJL5 V5G2P6 E0VW24 A0A0P4VWP1 T1IU24 A0A224XG59 A0A069DYE2 A0A0V0G3H5 J9K5L3 A0A2H8TY68 A0A210PF93 A0A2S2Q793 A0A2S2NPD4 A0A2R2MPW2
PDB
6MOL
E-value=2.58308e-56,
Score=557
Ontologies
GO
Topology
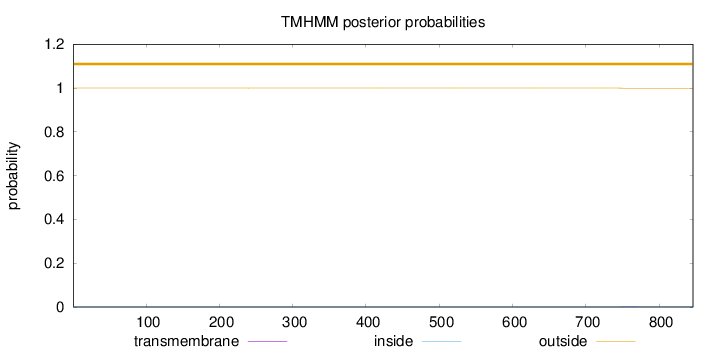
Length:
846
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01615
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00019
outside
1 - 846
Population Genetic Test Statistics
Pi
24.792887
Theta
22.725752
Tajima's D
-0.900639
CLR
1.303682
CSRT
0.154842257887106
Interpretation
Uncertain
