Gene
KWMTBOMO03610
Pre Gene Modal
BGIBMGA006325
Annotation
extra_sex_combs_[Bombyx_mori]
Full name
Polycomb protein EED
Location in the cell
Cytoplasmic Reliability : 1.271 Nuclear Reliability : 1.959
Sequence
CDS
ATGAATTTTTCTGAAAATGAAGCTGATGACACTTCAAGTGTTGATAGTACTTCAAACACAGACAATACGTCCCGTAGTGAGACTCCTACCAACTCAAGAGTGAAAAAACGTCGTCGCGGCAAAAAGAAAACCACTAAGCCAGTTAAACCTCCATACAAGTTCAATTGCAGTGCTAAAGAAGATCATGGTCAACCACTATTTGGATGCCAGTTCAATCATCATCTCGGTGAAGGAGAACCATCAGTATTTGCAGTAGTCGGTAGTAACAGAGTCTCAATATATGAGTGCCCTGAATCTGGAGGCTTCAAATTCTTGCAGTGCTATGCTGATCCTGATGTTGATGAAACATTTTACACATGCGCTTGGTCTTACGAAGAAGACACGATGCTTCCGCTCCTAGCCGTCGCCGGGTCACGCGGTATCATCCGTATCTTCCACCCCGCCACGCAGACGTGCATCAAACACTACGTGGGGCACGGACACGCCATCAACGAGGTTAAGTTCCATCCTCGAGACCCGAATCTACTTCTGTCGGCCAGCAAGGACCACGCTCTGCGGCTTTGGAACATTATGACTGACGTTTGCATAGCCATCTTCGGCGGGGTCGAGGGCCACAGAGACGAAGTGCTCAGCGCTGACTTCGATCTAAAAGGAGAAAGGATAATGTCCTGCGGGATGGACCACTCCCTGAAACTGTGGAGGTTGGACAAGCCCTCGATGAATGAGGCCATCAAACAGAGCTATAATTTCAATCCGCACAGAGCGCTACGTCCCTTCAATTCCTTGAAGGAACATTTTCCTGATTTTTCAACTAGAGATATACATAGGAATTACGTTGACTGCGTCAGGTGGATGGGAGATCTAATTCTTTCTAAATCGTGTGAAAACGCTATAATCTGCTGGAAGCCGGGCCGACTTGAGGACACGGAGCTGCGGCCCGGGGACAACTCCGTCACCATGGTGCATCGCTTCGACTACAAAGAGTGTGAAATATGGTTTATACGATTCGCTGTTGACTATAGTCAAAGGGTGATAGCTCTAGGTAATCAGTGCGGTAAGACCATGGTGTGGGAACTGGGGAACGTGGCTGGAGGCTCCCGAGTCTCGCAGCTCGTGCATCCGAGATGCGTTGCTGCGGTCAGACAGGTAACTCTCTCACGCAACGGCAAGATCCTACTGACGTGTTGCGACGACGGCACCATCTGGCGGTGGGACCGGGTCTCCGCCAGCAACTGA
Protein
MNFSENEADDTSSVDSTSNTDNTSRSETPTNSRVKKRRRGKKKTTKPVKPPYKFNCSAKEDHGQPLFGCQFNHHLGEGEPSVFAVVGSNRVSIYECPESGGFKFLQCYADPDVDETFYTCAWSYEEDTMLPLLAVAGSRGIIRIFHPATQTCIKHYVGHGHAINEVKFHPRDPNLLLSASKDHALRLWNIMTDVCIAIFGGVEGHRDEVLSADFDLKGERIMSCGMDHSLKLWRLDKPSMNEAIKQSYNFNPHRALRPFNSLKEHFPDFSTRDIHRNYVDCVRWMGDLILSKSCENAIICWKPGRLEDTELRPGDNSVTMVHRFDYKECEIWFIRFAVDYSQRVIALGNQCGKTMVWELGNVAGGSRVSQLVHPRCVAAVRQVTLSRNGKILLTCCDDGTIWRWDRVSASN
Summary
Description
Polycomb group (PcG) protein. Component of the PRC2/EED-EZH2 complex, which methylates 'Lys-9' and 'Lys-27' of histone H3, leading to transcriptional repression of the affected target gene (By similarity).
Subunit
Component of the PRC2/EED-EZH2 complex.
Similarity
Belongs to the WD repeat ESC family.
Keywords
Chromatin regulator
Complete proteome
Nucleus
Reference proteome
Repeat
Repressor
Transcription
Transcription regulation
WD repeat
Feature
chain Polycomb protein EED
Uniprot
E5RWX8
H9J9Y0
O16021
A0A2A4JTR0
A0A2W1BV52
A0A212F587
+ More
R4IT83 A0A194R7G9 A0A194QA22 A0A2H1WSP0 O16022 A0A2J7PDJ4 A0A067QSB7 A0A347ZJF6 A0A2P8ZKJ7 A0A347ZJF7 A0A1B6LV29 A0A1B6EUS2 U4UG71 N6U559 A0A139WM98 D6WCC2 A0A1W4WS66 J3JV55 A0A1W4WHA9 A0A1Y1LJT0 A0A1L7N283 E9ISN8 A0A0L7QWH6 A0A023F4K4 T1HXR8 A0A224XQR9 A0A069DT80 E0VN79 A0A154P6U4 A0A151WMC0 A0A0M8ZYT7 A0A026WPI2 A0A232FEF1 K7IRS7 A0A0J7P319 E2ADP6 A0A195E5K6 A0A195EVB7 A0A151IH66 F4WP90 A0A2A3ES72 A0A195B0U1 A0A158NR18 A0A1B0GKP6 E2BUS5 A0A0L7LE57 A0A0C9R285 T1J4Y1 A0A1B6EG92 E9H4Z8 A0A164R077 A0A0N8ATL3 A0A0P5J3H1 Q16G31 A0A182H2R0 A0A1Q3EWC0 A0A0P5JC02 H0ZRX6 A0A218UJ00 A0A151NM02 A0A1L1RP86 A0A2I0M127 A0A091FR44 A0A091FET9 A0A093C3F1 A0A091GTF5 A0A099ZGR5 A0A093SQ30 A0A091IU09 A0A091WN24 A0A091LWQ3 A0A091W017 A0A093GD39 A0A091NIQ8 A0A091J1R3 A0A0A0A822 A0A093DKT2 A0A093NXK1 A0A093F8D1 A0A091KNS1 A0A093JMQ8 A0A091QCZ8 Q5ZKH3 R0L1L4 G1NQR4 A0A1V4JR49 A0A0Q3PM19 U3JZ14 U3IZS5 F6TW64 J9K6S7 G3WKF2 A0A3Q0HLW3 G1KAU8
R4IT83 A0A194R7G9 A0A194QA22 A0A2H1WSP0 O16022 A0A2J7PDJ4 A0A067QSB7 A0A347ZJF6 A0A2P8ZKJ7 A0A347ZJF7 A0A1B6LV29 A0A1B6EUS2 U4UG71 N6U559 A0A139WM98 D6WCC2 A0A1W4WS66 J3JV55 A0A1W4WHA9 A0A1Y1LJT0 A0A1L7N283 E9ISN8 A0A0L7QWH6 A0A023F4K4 T1HXR8 A0A224XQR9 A0A069DT80 E0VN79 A0A154P6U4 A0A151WMC0 A0A0M8ZYT7 A0A026WPI2 A0A232FEF1 K7IRS7 A0A0J7P319 E2ADP6 A0A195E5K6 A0A195EVB7 A0A151IH66 F4WP90 A0A2A3ES72 A0A195B0U1 A0A158NR18 A0A1B0GKP6 E2BUS5 A0A0L7LE57 A0A0C9R285 T1J4Y1 A0A1B6EG92 E9H4Z8 A0A164R077 A0A0N8ATL3 A0A0P5J3H1 Q16G31 A0A182H2R0 A0A1Q3EWC0 A0A0P5JC02 H0ZRX6 A0A218UJ00 A0A151NM02 A0A1L1RP86 A0A2I0M127 A0A091FR44 A0A091FET9 A0A093C3F1 A0A091GTF5 A0A099ZGR5 A0A093SQ30 A0A091IU09 A0A091WN24 A0A091LWQ3 A0A091W017 A0A093GD39 A0A091NIQ8 A0A091J1R3 A0A0A0A822 A0A093DKT2 A0A093NXK1 A0A093F8D1 A0A091KNS1 A0A093JMQ8 A0A091QCZ8 Q5ZKH3 R0L1L4 G1NQR4 A0A1V4JR49 A0A0Q3PM19 U3JZ14 U3IZS5 F6TW64 J9K6S7 G3WKF2 A0A3Q0HLW3 G1KAU8
Pubmed
22485166
19121390
9343430
28756777
22118469
23814061
+ More
26354079 24845553 29403074 23537049 18362917 19820115 22516182 28004739 27956627 21282665 25474469 26334808 20566863 24508170 30249741 28648823 20075255 20798317 21719571 21347285 26227816 21292972 17510324 26483478 20360741 22293439 23371554 15642098 20838655 23749191 17495919 21709235
26354079 24845553 29403074 23537049 18362917 19820115 22516182 28004739 27956627 21282665 25474469 26334808 20566863 24508170 30249741 28648823 20075255 20798317 21719571 21347285 26227816 21292972 17510324 26483478 20360741 22293439 23371554 15642098 20838655 23749191 17495919 21709235
EMBL
AB607838
BAJ54072.1
BABH01026037
AF003603
AAC05331.1
NWSH01000685
+ More
PCG74790.1 KZ149893 PZC78938.1 AGBW02010219 OWR48902.1 JQ744271 AGC39037.1 KQ460647 KPJ13185.1 KQ459249 KPJ02373.1 ODYU01010734 SOQ56027.1 AF003604 AAC05332.1 NEVH01026386 PNF14398.1 KK853353 KDR08216.1 FX985724 BBA84462.1 PYGN01000028 PSN57027.1 FX985725 BBA84463.1 GEBQ01012439 JAT27538.1 GECZ01028024 JAS41745.1 KB632309 ERL92032.1 APGK01042227 KB741002 ENN75776.1 KQ971316 KYB29013.1 EEZ98792.2 BT127121 AEE62083.1 GEZM01053849 JAV73922.1 LC100105 BAW19570.1 GL765434 EFZ16293.1 KQ414713 KOC62955.1 GBBI01002510 JAC16202.1 ACPB03021030 GFTR01006085 JAW10341.1 GBGD01001591 JAC87298.1 DS235335 EEB14835.1 KQ434827 KZC07597.1 KQ982944 KYQ49014.1 KQ435794 KOX73880.1 KK107139 QOIP01000003 EZA57883.1 RLU24378.1 NNAY01000331 OXU29164.1 LBMM01000241 KMR03299.1 GL438820 EFN68408.1 KQ979608 KYN20475.1 KQ981965 KYN31834.1 KQ977637 KYN01135.1 GL888243 EGI64071.1 KZ288189 PBC34643.1 KQ976691 KYM77912.1 ADTU01023744 AJWK01030843 GL450746 EFN80548.1 JTDY01001482 KOB73767.1 GBYB01010209 JAG79976.1 JH431850 GEDC01000332 JAS36966.1 GL732592 EFX73235.1 LRGB01002285 KZS08223.1 GDIQ01244155 JAK07570.1 GDIQ01203666 JAK48059.1 CH478339 EAT33200.1 JXUM01003383 KQ560157 KXJ84133.1 GFDL01015444 JAV19601.1 GDIQ01227207 JAK24518.1 ABQF01027776 MUZQ01000270 OWK53704.1 AKHW03002570 KYO37818.1 AKCR02000051 PKK23378.1 KL447262 KFO71436.1 KK718917 KFO60070.1 KL452576 KFV06797.1 KL508517 KFO85685.1 KL892873 KGL80238.1 KL672094 KFW84699.1 KL218233 KFP03141.1 KL411053 KFR02901.1 KK510031 KFP62787.1 KK734465 KFR08365.1 KL215889 KFV67113.1 KK834501 KFP79027.1 KK501377 KFP14522.1 KL871085 KGL90221.1 KN126888 KFU93312.1 KL225030 KFW66695.1 KK388299 KFV52770.1 KK748189 KFP40935.1 KL206225 KFV80029.1 KK695283 KFQ23948.1 AJ720111 KB744949 EOA94172.1 LSYS01006642 OPJ74678.1 LMAW01002014 KQK82075.1 AGTO01010903 ADON01136942 ADON01136943 ADON01136944 ADON01136945 ADON01136946 ABLF02034383 AEFK01118901 AEFK01118902
PCG74790.1 KZ149893 PZC78938.1 AGBW02010219 OWR48902.1 JQ744271 AGC39037.1 KQ460647 KPJ13185.1 KQ459249 KPJ02373.1 ODYU01010734 SOQ56027.1 AF003604 AAC05332.1 NEVH01026386 PNF14398.1 KK853353 KDR08216.1 FX985724 BBA84462.1 PYGN01000028 PSN57027.1 FX985725 BBA84463.1 GEBQ01012439 JAT27538.1 GECZ01028024 JAS41745.1 KB632309 ERL92032.1 APGK01042227 KB741002 ENN75776.1 KQ971316 KYB29013.1 EEZ98792.2 BT127121 AEE62083.1 GEZM01053849 JAV73922.1 LC100105 BAW19570.1 GL765434 EFZ16293.1 KQ414713 KOC62955.1 GBBI01002510 JAC16202.1 ACPB03021030 GFTR01006085 JAW10341.1 GBGD01001591 JAC87298.1 DS235335 EEB14835.1 KQ434827 KZC07597.1 KQ982944 KYQ49014.1 KQ435794 KOX73880.1 KK107139 QOIP01000003 EZA57883.1 RLU24378.1 NNAY01000331 OXU29164.1 LBMM01000241 KMR03299.1 GL438820 EFN68408.1 KQ979608 KYN20475.1 KQ981965 KYN31834.1 KQ977637 KYN01135.1 GL888243 EGI64071.1 KZ288189 PBC34643.1 KQ976691 KYM77912.1 ADTU01023744 AJWK01030843 GL450746 EFN80548.1 JTDY01001482 KOB73767.1 GBYB01010209 JAG79976.1 JH431850 GEDC01000332 JAS36966.1 GL732592 EFX73235.1 LRGB01002285 KZS08223.1 GDIQ01244155 JAK07570.1 GDIQ01203666 JAK48059.1 CH478339 EAT33200.1 JXUM01003383 KQ560157 KXJ84133.1 GFDL01015444 JAV19601.1 GDIQ01227207 JAK24518.1 ABQF01027776 MUZQ01000270 OWK53704.1 AKHW03002570 KYO37818.1 AKCR02000051 PKK23378.1 KL447262 KFO71436.1 KK718917 KFO60070.1 KL452576 KFV06797.1 KL508517 KFO85685.1 KL892873 KGL80238.1 KL672094 KFW84699.1 KL218233 KFP03141.1 KL411053 KFR02901.1 KK510031 KFP62787.1 KK734465 KFR08365.1 KL215889 KFV67113.1 KK834501 KFP79027.1 KK501377 KFP14522.1 KL871085 KGL90221.1 KN126888 KFU93312.1 KL225030 KFW66695.1 KK388299 KFV52770.1 KK748189 KFP40935.1 KL206225 KFV80029.1 KK695283 KFQ23948.1 AJ720111 KB744949 EOA94172.1 LSYS01006642 OPJ74678.1 LMAW01002014 KQK82075.1 AGTO01010903 ADON01136942 ADON01136943 ADON01136944 ADON01136945 ADON01136946 ABLF02034383 AEFK01118901 AEFK01118902
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000053268
UP000235965
+ More
UP000027135 UP000245037 UP000030742 UP000019118 UP000007266 UP000192223 UP000053825 UP000015103 UP000009046 UP000076502 UP000075809 UP000053105 UP000053097 UP000279307 UP000215335 UP000002358 UP000036403 UP000000311 UP000078492 UP000078541 UP000078542 UP000007755 UP000242457 UP000078540 UP000005205 UP000092461 UP000008237 UP000037510 UP000000305 UP000076858 UP000008820 UP000069940 UP000249989 UP000007754 UP000197619 UP000050525 UP000053872 UP000053760 UP000052976 UP000053641 UP000053258 UP000054308 UP000053283 UP000053605 UP000053875 UP000053119 UP000053858 UP000054081 UP000053584 UP000000539 UP000001645 UP000190648 UP000051836 UP000016665 UP000016666 UP000002280 UP000007819 UP000007648 UP000189705 UP000001646
UP000027135 UP000245037 UP000030742 UP000019118 UP000007266 UP000192223 UP000053825 UP000015103 UP000009046 UP000076502 UP000075809 UP000053105 UP000053097 UP000279307 UP000215335 UP000002358 UP000036403 UP000000311 UP000078492 UP000078541 UP000078542 UP000007755 UP000242457 UP000078540 UP000005205 UP000092461 UP000008237 UP000037510 UP000000305 UP000076858 UP000008820 UP000069940 UP000249989 UP000007754 UP000197619 UP000050525 UP000053872 UP000053760 UP000052976 UP000053641 UP000053258 UP000054308 UP000053283 UP000053605 UP000053875 UP000053119 UP000053858 UP000054081 UP000053584 UP000000539 UP000001645 UP000190648 UP000051836 UP000016665 UP000016666 UP000002280 UP000007819 UP000007648 UP000189705 UP000001646
Interpro
Gene 3D
ProteinModelPortal
E5RWX8
H9J9Y0
O16021
A0A2A4JTR0
A0A2W1BV52
A0A212F587
+ More
R4IT83 A0A194R7G9 A0A194QA22 A0A2H1WSP0 O16022 A0A2J7PDJ4 A0A067QSB7 A0A347ZJF6 A0A2P8ZKJ7 A0A347ZJF7 A0A1B6LV29 A0A1B6EUS2 U4UG71 N6U559 A0A139WM98 D6WCC2 A0A1W4WS66 J3JV55 A0A1W4WHA9 A0A1Y1LJT0 A0A1L7N283 E9ISN8 A0A0L7QWH6 A0A023F4K4 T1HXR8 A0A224XQR9 A0A069DT80 E0VN79 A0A154P6U4 A0A151WMC0 A0A0M8ZYT7 A0A026WPI2 A0A232FEF1 K7IRS7 A0A0J7P319 E2ADP6 A0A195E5K6 A0A195EVB7 A0A151IH66 F4WP90 A0A2A3ES72 A0A195B0U1 A0A158NR18 A0A1B0GKP6 E2BUS5 A0A0L7LE57 A0A0C9R285 T1J4Y1 A0A1B6EG92 E9H4Z8 A0A164R077 A0A0N8ATL3 A0A0P5J3H1 Q16G31 A0A182H2R0 A0A1Q3EWC0 A0A0P5JC02 H0ZRX6 A0A218UJ00 A0A151NM02 A0A1L1RP86 A0A2I0M127 A0A091FR44 A0A091FET9 A0A093C3F1 A0A091GTF5 A0A099ZGR5 A0A093SQ30 A0A091IU09 A0A091WN24 A0A091LWQ3 A0A091W017 A0A093GD39 A0A091NIQ8 A0A091J1R3 A0A0A0A822 A0A093DKT2 A0A093NXK1 A0A093F8D1 A0A091KNS1 A0A093JMQ8 A0A091QCZ8 Q5ZKH3 R0L1L4 G1NQR4 A0A1V4JR49 A0A0Q3PM19 U3JZ14 U3IZS5 F6TW64 J9K6S7 G3WKF2 A0A3Q0HLW3 G1KAU8
R4IT83 A0A194R7G9 A0A194QA22 A0A2H1WSP0 O16022 A0A2J7PDJ4 A0A067QSB7 A0A347ZJF6 A0A2P8ZKJ7 A0A347ZJF7 A0A1B6LV29 A0A1B6EUS2 U4UG71 N6U559 A0A139WM98 D6WCC2 A0A1W4WS66 J3JV55 A0A1W4WHA9 A0A1Y1LJT0 A0A1L7N283 E9ISN8 A0A0L7QWH6 A0A023F4K4 T1HXR8 A0A224XQR9 A0A069DT80 E0VN79 A0A154P6U4 A0A151WMC0 A0A0M8ZYT7 A0A026WPI2 A0A232FEF1 K7IRS7 A0A0J7P319 E2ADP6 A0A195E5K6 A0A195EVB7 A0A151IH66 F4WP90 A0A2A3ES72 A0A195B0U1 A0A158NR18 A0A1B0GKP6 E2BUS5 A0A0L7LE57 A0A0C9R285 T1J4Y1 A0A1B6EG92 E9H4Z8 A0A164R077 A0A0N8ATL3 A0A0P5J3H1 Q16G31 A0A182H2R0 A0A1Q3EWC0 A0A0P5JC02 H0ZRX6 A0A218UJ00 A0A151NM02 A0A1L1RP86 A0A2I0M127 A0A091FR44 A0A091FET9 A0A093C3F1 A0A091GTF5 A0A099ZGR5 A0A093SQ30 A0A091IU09 A0A091WN24 A0A091LWQ3 A0A091W017 A0A093GD39 A0A091NIQ8 A0A091J1R3 A0A0A0A822 A0A093DKT2 A0A093NXK1 A0A093F8D1 A0A091KNS1 A0A093JMQ8 A0A091QCZ8 Q5ZKH3 R0L1L4 G1NQR4 A0A1V4JR49 A0A0Q3PM19 U3JZ14 U3IZS5 F6TW64 J9K6S7 G3WKF2 A0A3Q0HLW3 G1KAU8
PDB
5H13
E-value=4.4319e-146,
Score=1328
Ontologies
GO
PANTHER
Topology
Subcellular location
Nucleus
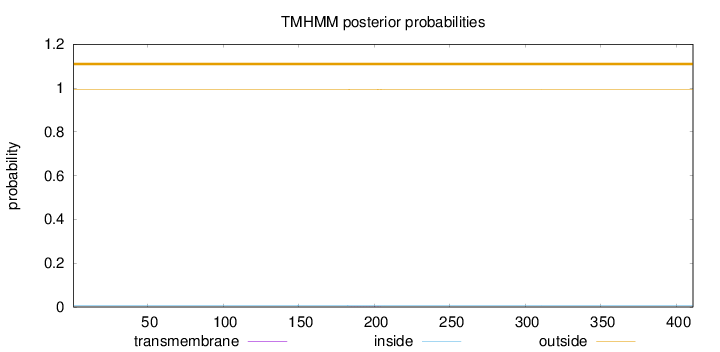
Length:
411
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00987999999999999
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00719
outside
1 - 411
Population Genetic Test Statistics
Pi
32.082621
Theta
25.877984
Tajima's D
-0.61454
CLR
0.502427
CSRT
0.219189040547973
Interpretation
Uncertain
