Gene
KWMTBOMO03608 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA006272
Annotation
37-kDa_protease_precursor_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 2.739
Sequence
CDS
ATGGTAATAATTTCAGAATGCGGCATAGCACGCACTCGTCGGCGCATCGTGGGCGGATATGAAACGAAAGAGACGGAGTACCCCTGGATGGCCGCTCTTTTGTACGGCGGAAGATTCTATTGTGGTGGTGCACTTATCAGTGATCTGTACGTTTTGACAGCTGCTCATTGTACTTCAGGATTCCGCAAGGAACGGATTACAGTTCGGTTCTTGGAGCACGATCGTTCTAAAGTAAACGAAACTAAAACGATAGACAGAAAGGTGTCTGACATCATTCGTCATCTGCGGTATAATCCCGGAACTTACGACAGTGATATCGCCCTTTTAAAACTAGCTGAGAGGGTAGACCTCAGCAGTGCATTGAAGCGAGTTCGCAGTGAAGGAGACAACGGCACTGCCACGGATGACGACAAGGACGTCGGGCTAAGACCGGTCTGTTTACCCAGTTCTGGACTCTCCTATAACAATTACACGGGTGTTGTCACAGGCTGGGGAACTACAGAGGAAGGTGGCTCTGTATCCAATGCATTACAGGAGGTGAAAGTACCGATTGTGACAAATGAAGAATGTCGTAAAGGCTACGGTGATCGGATAACAGATAATATGATTTGCGCTGGAGAGCCAGAGGGCGGCCGTGACGCTTGTCAGGGAGACTCGGGTGGACCGATGCATGTTCTTGAAATTGAGACATCAAAATACTCTGAAGTCGGTGTCGTGTCTTGGGGCGAAGGGTGCGCGCGACCAAACAAACCAGGCGTTTATACACGTGTCAATCGATACCTCACTTGGATTAAACAGAACACTCGCGATGCCTGCAATTGTCAATAA
Protein
MVIISECGIARTRRRIVGGYETKETEYPWMAALLYGGRFYCGGALISDLYVLTAAHCTSGFRKERITVRFLEHDRSKVNETKTIDRKVSDIIRHLRYNPGTYDSDIALLKLAERVDLSSALKRVRSEGDNGTATDDDKDVGLRPVCLPSSGLSYNNYTGVVTGWGTTEEGGSVSNALQEVKVPIVTNEECRKGYGDRITDNMICAGEPEGGRDACQGDSGGPMHVLEIETSKYSEVGVVSWGEGCARPNKPGVYTRVNRYLTWIKQNTRDACNCQ
Summary
Similarity
Belongs to the peptidase S1 family.
Uniprot
B3Y578
H9J9S9
A0A2H1WSI3
A0A2A4JSW9
I4DJ19
I4DM81
+ More
A0A194QA26 A0A194R692 A0A2W1C068 A0A1B6E201 A0A068F7A2 A0A1B6DH02 A0A1I8N4Z3 A0A1I8NVF3 E0VW09 B3NN20 B4QGL3 B4MS13 Q9W2C8 B4P8E6 A0A1W4UN83 A0A2P8ZM46 B4I7W6 A0A0L0CM55 U5EYE8 D6W9P1 B4J6A3 A0A067QL67 B4GHF3 Q28XQ9 A0A0K8W9A7 A0A034WBK0 A0A1A9Y7Y7 A0A3B0JPZ2 A0A0A1WGA3 A0A0A1WT31 A0A2J7Q474 W8BMT4 A0A0Q9XLY6 B3MJ08 A0A1W4X406 A0A1W4WUG0 A0A087ZRI1 Q16G07 A0A2A3E772 A0A1J1IYI4 A0A1B0ARI0 A0A0M4EIU8 A0A023F2N0 A0A084WT80 B0WAI6 Q28XR0 A0A023ENP9 A0A182QYZ9 A0A1A9ZRH0 A0A1V1G177 B4GHF4 Q16G08 A0A3B0J2K7 A0A1A9VEE8 A0A2S2Q7D4 W8C9P6 A0A034VQ73 A0A1W4UYQ3 A0A182N9T7 A0A0A1XSC5 A0A154PBY1 A0A0L0CMI8 B4J6A2 A0A0K8WI64 J9K442 A0A1S4G2A2 B3NN19 A0A182PQS9 B4P8E7 A0A182MI07 E2BDT9 Q9I7V4 B4I7W5 A0A195EJ73 W5JJ63 A0A0M5IZN1 B4MS12 A0A2H8TLU6 B4QGL2 A0A182HYU6 A0A2C9H896 Q7Q9W5 A0A195D4U7 A0A182KZD5 A0A195F5K4 A0A1A9X3F3 A0A146MG71 A0A182JXD4 A0A232ELK8 F4X0S0 K7J298 Q28XQ4 A0A158P398 A0A182Y8L4
A0A194QA26 A0A194R692 A0A2W1C068 A0A1B6E201 A0A068F7A2 A0A1B6DH02 A0A1I8N4Z3 A0A1I8NVF3 E0VW09 B3NN20 B4QGL3 B4MS13 Q9W2C8 B4P8E6 A0A1W4UN83 A0A2P8ZM46 B4I7W6 A0A0L0CM55 U5EYE8 D6W9P1 B4J6A3 A0A067QL67 B4GHF3 Q28XQ9 A0A0K8W9A7 A0A034WBK0 A0A1A9Y7Y7 A0A3B0JPZ2 A0A0A1WGA3 A0A0A1WT31 A0A2J7Q474 W8BMT4 A0A0Q9XLY6 B3MJ08 A0A1W4X406 A0A1W4WUG0 A0A087ZRI1 Q16G07 A0A2A3E772 A0A1J1IYI4 A0A1B0ARI0 A0A0M4EIU8 A0A023F2N0 A0A084WT80 B0WAI6 Q28XR0 A0A023ENP9 A0A182QYZ9 A0A1A9ZRH0 A0A1V1G177 B4GHF4 Q16G08 A0A3B0J2K7 A0A1A9VEE8 A0A2S2Q7D4 W8C9P6 A0A034VQ73 A0A1W4UYQ3 A0A182N9T7 A0A0A1XSC5 A0A154PBY1 A0A0L0CMI8 B4J6A2 A0A0K8WI64 J9K442 A0A1S4G2A2 B3NN19 A0A182PQS9 B4P8E7 A0A182MI07 E2BDT9 Q9I7V4 B4I7W5 A0A195EJ73 W5JJ63 A0A0M5IZN1 B4MS12 A0A2H8TLU6 B4QGL2 A0A182HYU6 A0A2C9H896 Q7Q9W5 A0A195D4U7 A0A182KZD5 A0A195F5K4 A0A1A9X3F3 A0A146MG71 A0A182JXD4 A0A232ELK8 F4X0S0 K7J298 Q28XQ4 A0A158P398 A0A182Y8L4
Pubmed
19114104
19121390
22651552
26354079
28756777
24952583
+ More
25315136 20566863 17994087 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 29403074 26108605 18362917 19820115 24845553 15632085 25348373 25830018 24495485 17510324 25474469 24438588 24945155 28410430 20798317 20920257 23761445 12364791 14747013 17210077 20966253 26823975 28648823 21719571 20075255 21347285 25244985
25315136 20566863 17994087 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 29403074 26108605 18362917 19820115 24845553 15632085 25348373 25830018 24495485 17510324 25474469 24438588 24945155 28410430 20798317 20920257 23761445 12364791 14747013 17210077 20966253 26823975 28648823 21719571 20075255 21347285 25244985
EMBL
AB434653
BAG68694.1
BABH01026032
ODYU01010734
SOQ56030.1
NWSH01000685
+ More
PCG74788.1 AK401287 BAM17909.1 AK402399 BAM19021.1 KQ459249 KPJ02378.1 KQ460647 KPJ13187.1 KZ149893 PZC78907.1 GEDC01005335 GEDC01000236 JAS31963.1 JAS37062.1 KJ512113 KJ512117 AID60336.1 AID60340.1 GEDC01012317 GEDC01010410 JAS24981.1 JAS26888.1 DS235816 EEB17565.1 CH954179 EDV55514.1 CM000362 CM002911 EDX08132.1 KMY95663.1 CH963850 EDW74902.1 AE013599 AY052010 AAF46764.2 AAK93434.1 CM000158 EDW91186.1 PYGN01000018 PSN57576.1 CH480824 EDW56691.1 JRES01000195 KNC33428.1 GANO01000344 JAB59527.1 KQ971312 EEZ99266.1 CH916367 EDW00876.1 KK853216 KDR09773.1 CH479183 EDW35923.1 CM000071 EAL26257.2 GDHF01004652 JAI47662.1 GAKP01007799 JAC51153.1 OUUW01000001 SPP75416.1 GBXI01016581 JAC97710.1 GBXI01012709 JAD01583.1 NEVH01018390 PNF23369.1 GAMC01006453 JAC00103.1 CH933808 KRG05032.1 CH902619 EDV37074.2 CH478352 EAT33171.1 KZ288360 PBC27019.1 CVRI01000064 CRL05277.1 JXJN01002447 CP012524 ALC40949.1 ALC42785.1 GBBI01003132 JAC15580.1 ATLV01026865 KE525420 KFB53424.1 DS231872 EDS41409.1 EAL26256.2 GAPW01002546 JAC11052.1 AXCN02001200 FX985324 BAX07337.1 EDW35924.1 EAT33170.1 SPP75417.1 GGMS01004462 MBY73665.1 GAMC01006451 JAC00105.1 GAKP01014333 JAC44619.1 GBXI01000849 JAD13443.1 KQ434869 KZC09429.1 KNC33427.1 EDW00875.1 GDHF01001547 JAI50767.1 ABLF02030882 EDV55513.1 EDW91187.1 AXCM01002956 GL447689 EFN86145.1 BT125606 AAG22193.1 ADM26844.1 EDW56690.1 KQ978801 KYN28211.1 ADMH02001350 ETN62920.1 ALC40948.1 ALC42784.1 EDW74901.1 GFXV01003290 MBW15095.1 EDX08131.1 KMY95662.1 APCN01005409 AAAB01008898 EAA09293.5 KQ976842 KYN07908.1 KQ981805 KYN35462.1 GDHC01000394 JAQ18235.1 NNAY01003532 OXU19246.1 GL888499 EGI59947.1 AAZX01006360 EAL26262.3 ADTU01007949
PCG74788.1 AK401287 BAM17909.1 AK402399 BAM19021.1 KQ459249 KPJ02378.1 KQ460647 KPJ13187.1 KZ149893 PZC78907.1 GEDC01005335 GEDC01000236 JAS31963.1 JAS37062.1 KJ512113 KJ512117 AID60336.1 AID60340.1 GEDC01012317 GEDC01010410 JAS24981.1 JAS26888.1 DS235816 EEB17565.1 CH954179 EDV55514.1 CM000362 CM002911 EDX08132.1 KMY95663.1 CH963850 EDW74902.1 AE013599 AY052010 AAF46764.2 AAK93434.1 CM000158 EDW91186.1 PYGN01000018 PSN57576.1 CH480824 EDW56691.1 JRES01000195 KNC33428.1 GANO01000344 JAB59527.1 KQ971312 EEZ99266.1 CH916367 EDW00876.1 KK853216 KDR09773.1 CH479183 EDW35923.1 CM000071 EAL26257.2 GDHF01004652 JAI47662.1 GAKP01007799 JAC51153.1 OUUW01000001 SPP75416.1 GBXI01016581 JAC97710.1 GBXI01012709 JAD01583.1 NEVH01018390 PNF23369.1 GAMC01006453 JAC00103.1 CH933808 KRG05032.1 CH902619 EDV37074.2 CH478352 EAT33171.1 KZ288360 PBC27019.1 CVRI01000064 CRL05277.1 JXJN01002447 CP012524 ALC40949.1 ALC42785.1 GBBI01003132 JAC15580.1 ATLV01026865 KE525420 KFB53424.1 DS231872 EDS41409.1 EAL26256.2 GAPW01002546 JAC11052.1 AXCN02001200 FX985324 BAX07337.1 EDW35924.1 EAT33170.1 SPP75417.1 GGMS01004462 MBY73665.1 GAMC01006451 JAC00105.1 GAKP01014333 JAC44619.1 GBXI01000849 JAD13443.1 KQ434869 KZC09429.1 KNC33427.1 EDW00875.1 GDHF01001547 JAI50767.1 ABLF02030882 EDV55513.1 EDW91187.1 AXCM01002956 GL447689 EFN86145.1 BT125606 AAG22193.1 ADM26844.1 EDW56690.1 KQ978801 KYN28211.1 ADMH02001350 ETN62920.1 ALC40948.1 ALC42784.1 EDW74901.1 GFXV01003290 MBW15095.1 EDX08131.1 KMY95662.1 APCN01005409 AAAB01008898 EAA09293.5 KQ976842 KYN07908.1 KQ981805 KYN35462.1 GDHC01000394 JAQ18235.1 NNAY01003532 OXU19246.1 GL888499 EGI59947.1 AAZX01006360 EAL26262.3 ADTU01007949
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000095301
UP000095300
+ More
UP000009046 UP000008711 UP000000304 UP000007798 UP000000803 UP000002282 UP000192221 UP000245037 UP000001292 UP000037069 UP000007266 UP000001070 UP000027135 UP000008744 UP000001819 UP000092443 UP000268350 UP000235965 UP000009192 UP000007801 UP000192223 UP000005203 UP000008820 UP000242457 UP000183832 UP000092460 UP000092553 UP000030765 UP000002320 UP000075886 UP000092445 UP000078200 UP000075884 UP000076502 UP000007819 UP000075885 UP000075883 UP000008237 UP000078492 UP000000673 UP000075840 UP000076407 UP000007062 UP000078542 UP000075882 UP000078541 UP000091820 UP000075881 UP000215335 UP000007755 UP000002358 UP000005205 UP000076408
UP000009046 UP000008711 UP000000304 UP000007798 UP000000803 UP000002282 UP000192221 UP000245037 UP000001292 UP000037069 UP000007266 UP000001070 UP000027135 UP000008744 UP000001819 UP000092443 UP000268350 UP000235965 UP000009192 UP000007801 UP000192223 UP000005203 UP000008820 UP000242457 UP000183832 UP000092460 UP000092553 UP000030765 UP000002320 UP000075886 UP000092445 UP000078200 UP000075884 UP000076502 UP000007819 UP000075885 UP000075883 UP000008237 UP000078492 UP000000673 UP000075840 UP000076407 UP000007062 UP000078542 UP000075882 UP000078541 UP000091820 UP000075881 UP000215335 UP000007755 UP000002358 UP000005205 UP000076408
Pfam
PF00089 Trypsin
Interpro
SUPFAM
SSF50494
SSF50494
CDD
ProteinModelPortal
B3Y578
H9J9S9
A0A2H1WSI3
A0A2A4JSW9
I4DJ19
I4DM81
+ More
A0A194QA26 A0A194R692 A0A2W1C068 A0A1B6E201 A0A068F7A2 A0A1B6DH02 A0A1I8N4Z3 A0A1I8NVF3 E0VW09 B3NN20 B4QGL3 B4MS13 Q9W2C8 B4P8E6 A0A1W4UN83 A0A2P8ZM46 B4I7W6 A0A0L0CM55 U5EYE8 D6W9P1 B4J6A3 A0A067QL67 B4GHF3 Q28XQ9 A0A0K8W9A7 A0A034WBK0 A0A1A9Y7Y7 A0A3B0JPZ2 A0A0A1WGA3 A0A0A1WT31 A0A2J7Q474 W8BMT4 A0A0Q9XLY6 B3MJ08 A0A1W4X406 A0A1W4WUG0 A0A087ZRI1 Q16G07 A0A2A3E772 A0A1J1IYI4 A0A1B0ARI0 A0A0M4EIU8 A0A023F2N0 A0A084WT80 B0WAI6 Q28XR0 A0A023ENP9 A0A182QYZ9 A0A1A9ZRH0 A0A1V1G177 B4GHF4 Q16G08 A0A3B0J2K7 A0A1A9VEE8 A0A2S2Q7D4 W8C9P6 A0A034VQ73 A0A1W4UYQ3 A0A182N9T7 A0A0A1XSC5 A0A154PBY1 A0A0L0CMI8 B4J6A2 A0A0K8WI64 J9K442 A0A1S4G2A2 B3NN19 A0A182PQS9 B4P8E7 A0A182MI07 E2BDT9 Q9I7V4 B4I7W5 A0A195EJ73 W5JJ63 A0A0M5IZN1 B4MS12 A0A2H8TLU6 B4QGL2 A0A182HYU6 A0A2C9H896 Q7Q9W5 A0A195D4U7 A0A182KZD5 A0A195F5K4 A0A1A9X3F3 A0A146MG71 A0A182JXD4 A0A232ELK8 F4X0S0 K7J298 Q28XQ4 A0A158P398 A0A182Y8L4
A0A194QA26 A0A194R692 A0A2W1C068 A0A1B6E201 A0A068F7A2 A0A1B6DH02 A0A1I8N4Z3 A0A1I8NVF3 E0VW09 B3NN20 B4QGL3 B4MS13 Q9W2C8 B4P8E6 A0A1W4UN83 A0A2P8ZM46 B4I7W6 A0A0L0CM55 U5EYE8 D6W9P1 B4J6A3 A0A067QL67 B4GHF3 Q28XQ9 A0A0K8W9A7 A0A034WBK0 A0A1A9Y7Y7 A0A3B0JPZ2 A0A0A1WGA3 A0A0A1WT31 A0A2J7Q474 W8BMT4 A0A0Q9XLY6 B3MJ08 A0A1W4X406 A0A1W4WUG0 A0A087ZRI1 Q16G07 A0A2A3E772 A0A1J1IYI4 A0A1B0ARI0 A0A0M4EIU8 A0A023F2N0 A0A084WT80 B0WAI6 Q28XR0 A0A023ENP9 A0A182QYZ9 A0A1A9ZRH0 A0A1V1G177 B4GHF4 Q16G08 A0A3B0J2K7 A0A1A9VEE8 A0A2S2Q7D4 W8C9P6 A0A034VQ73 A0A1W4UYQ3 A0A182N9T7 A0A0A1XSC5 A0A154PBY1 A0A0L0CMI8 B4J6A2 A0A0K8WI64 J9K442 A0A1S4G2A2 B3NN19 A0A182PQS9 B4P8E7 A0A182MI07 E2BDT9 Q9I7V4 B4I7W5 A0A195EJ73 W5JJ63 A0A0M5IZN1 B4MS12 A0A2H8TLU6 B4QGL2 A0A182HYU6 A0A2C9H896 Q7Q9W5 A0A195D4U7 A0A182KZD5 A0A195F5K4 A0A1A9X3F3 A0A146MG71 A0A182JXD4 A0A232ELK8 F4X0S0 K7J298 Q28XQ4 A0A158P398 A0A182Y8L4
PDB
6O1G
E-value=1.19219e-38,
Score=399
Ontologies
GO
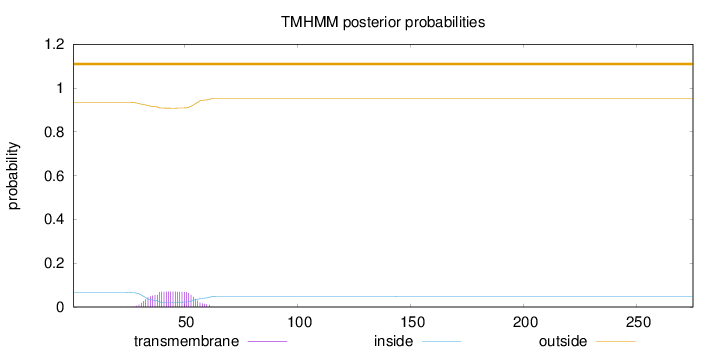
Topology
Length:
275
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.56803
Exp number, first 60 AAs:
1.55655
Total prob of N-in:
0.06599
outside
1 - 275
Population Genetic Test Statistics
Pi
28.522411
Theta
28.921111
Tajima's D
0.037593
CLR
2.310164
CSRT
0.380130993450327
Interpretation
Uncertain
