Pre Gene Modal
BGIBMGA006273
Annotation
PREDICTED:_glucose_transporter_type_1-like_isoform_X1_[Amyelois_transitella]
Full name
Glucose transporter type 1
Location in the cell
PlasmaMembrane Reliability : 4.966
Sequence
CDS
ATGTCGAGCGGTGGTACAACGGGGGCACTGCTGTACGCGGTGAGCGCGGCCGTCTTGGGAATGCTACAATTCGGCTTTAACACCGGCGTCATTAACGCCCCGAGAAAATTCATAGAGAACTTCGTACGTGATAATTACGAAGCGAAGCCGTCGCTCATTTTTAGTGTTATCGTCAGCATATTCGCCGTAGGAGGCATGATTGGATGTCCACTTGCCAGTTGGATGCTCGATACTCAGGGACGAAAGAAAGCCTTGCTCTATAACGCAGCATTCGGTATTATTGGAGCAGCTTTTATGGGTTTCAGTAAATTGGCGACTTCAGTTGAAATGCTCATCATTGGTAGATTCCTGATTGGAATCAACTGTGGCTTCGCAACGACGGCCTCGCCTACATACGTCTCCGAGATAGCACCGGTAAGAATGCGCGGCGCTTTCGGCACCGTAAACCAATTAGCTGTAGGATTCGGCATGACCTGTGGCCAGATTTTGGGTATTGATGTGATTTTGGGTAGCGACGAAGGCTGGCCGCTGTTGCTCGGATTAGCGGTCGTTCCGGCTGCCATTCAATTAATTATGCTACCGTTCGCGCCCGAATCACCAAGGTATTTGTTGCTATGCGCCCACGACGAAGAGAAAGCGAGAGTGGCCTTGACTAAAATTCGAGGCAGCAGCGATGTAGAAAACGAAATTAACGATATGCACCTTGAAGATCAAGCGGCCAGAGAAGAGCAGAAGTTCTCCGTGAAGGATTTGTTCACGACCCCAGCCCTAAGGATACCCCTCGTGATTGGTATCGTGATGCATTTATCCCAACAGTTGGGCGGAATCAACGCGGTATTGTACTATTCCTCCACGATATTTGTAAAAGCCGGTTTGTCTGAAGAAGACGCCCGGTTAGCTGTGATCGGTGTGGGCAGCATTCTTTTCGTGATGGCCCTCGTGTCGATTCCTCTGATGGACCGACTCGGCCGACGGACTCTGCAGCTGTGCGGTCTCGGCGGCATGACCGCATTCTCTATACTGATGACGATCGCGTTCTTCACGTACGAGAACAACACGACGATGAGTATATTCGCCGTGATATTCACATTCTTGTACGTCGCGTTCTTCGGTGTCGGACCTAGTTCGATCCCGTGGATGATCCTGTCGGAGTTGTTTGGTCAGGGAGCCAGGAGCGCGGCCGTCAGCGTCGGCGCTCTCGTTAACTGGTTCGCGAACTTCGTCGTGGGTCTAACGTTCATTCCTATGTCGGAAGCATTGGGGAACTACGTTTTCATACCGTACACGATCCTACTGATTATATTCTTCGTGTTCACGTATTTGAAGCTGCCCGAGACTAAGAACAGAACGATCGAGGAGGTGACGGCTCTCTTCAAGAAGTAG
Protein
MSSGGTTGALLYAVSAAVLGMLQFGFNTGVINAPRKFIENFVRDNYEAKPSLIFSVIVSIFAVGGMIGCPLASWMLDTQGRKKALLYNAAFGIIGAAFMGFSKLATSVEMLIIGRFLIGINCGFATTASPTYVSEIAPVRMRGAFGTVNQLAVGFGMTCGQILGIDVILGSDEGWPLLLGLAVVPAAIQLIMLPFAPESPRYLLLCAHDEEKARVALTKIRGSSDVENEINDMHLEDQAAREEQKFSVKDLFTTPALRIPLVIGIVMHLSQQLGGINAVLYYSSTIFVKAGLSEEDARLAVIGVGSILFVMALVSIPLMDRLGRRTLQLCGLGGMTAFSILMTIAFFTYENNTTMSIFAVIFTFLYVAFFGVGPSSIPWMILSELFGQGARSAAVSVGALVNWFANFVVGLTFIPMSEALGNYVFIPYTILLIIFFVFTYLKLPETKNRTIEEVTALFKK
Summary
Description
Facilitative glucose transporter.
Similarity
Belongs to the major facilitator superfamily. Sugar transporter (TC 2.A.1.1) family.
Belongs to the major facilitator superfamily. Sugar transporter (TC 2.A.1.1) family. Glucose transporter subfamily.
Belongs to the major facilitator superfamily. Sugar transporter (TC 2.A.1.1) family. Glucose transporter subfamily.
Keywords
Alternative splicing
Complete proteome
Glycoprotein
Membrane
Reference proteome
Signal
Sugar transport
Transmembrane
Transmembrane helix
Transport
Feature
chain Glucose transporter type 1
splice variant In isoform T.
splice variant In isoform T.
Uniprot
H9J9T0
A0A2W1BX91
A0A2A4JT42
A0A2A4JRX3
A0A2A4JSN4
A0A2H1WSK2
+ More
A0A0L7LE13 A0A194R6P4 A0A194QBL1 A0A212F4K9 Q8IRI6-10 Q8IRI6-2 A0A1L8DII3 T1P8T0 Q8IRI6-3 A0A2M4AR56 Q8IRI6-11 X2JBV9 W8AU68 A0A2M3Z9E3 W5JAR8 A0A2M4CJA9 A0A023EU33 A0A0S0WFC8 Q8IRI6-17 V9I6H3 A0A1Q3FWN3 A0A2A3EL61 Q17EP5 A0A2C9GPM1 A0A1L8DIT4 A0A1A9WPU6 X2JG36 Q8IRI6-16 Q7QJ79 Q8IRI6-12 A0A0J7KM10 Q8IRI6-14 Q8IRI6-6 Q8IRI6-15 A0A3L8DNS7 A0A026W3E5 A0A3B0JWX8 A0A1W4VUA3 A0A1Q3FX51 A0A2M4CUX3 B4IXB5 A0A1B6CWR9 B4QLH3 A0A087ZQD7 A0A1B6CTI4 A0A0L7R9B5 Q7PF87 A0A1I8MN39 A0A1I8PSE7 W0FVJ1 A0A1B6CQV5 A0A1B6DAY7 Q8IRI6-13 A0A023F5Q6 A0A1Z1XG22 T1IE15 B4HVL7 A0A0N8BMS5 A0A154PKQ2 A0A0N8EAH6 A0A146KMD7 A0A158NNR4 F4WV30 A0A164SYQ9 A0A0V0G868 A0A224XP36 A0A1B6CQC8 A0A1B6DC10 A0A069DV87 E9J9N5 A0A1B6EFA6 A0A1B6EAN5 B4KWH4 A0A146LVD3 A0A0Q9WIL2 A0A195BS38 A0A1B6DH78 A0A0P4VMS5 A0A1B6FH55 A0A1B6C2B8 A0A0K2UG24 A0A195EX69 A0A151WSZ8 A0A0P5C463 A0A2R5L7U2 A0A1B6LRT4 A0A0A8J8K9 A0A1B6L7Q0 A0A1B6J8Y5 A0A1B6MM35 A0A151IRI1 A0A0P5C448 A0A0P5C415 E2C2M2
A0A0L7LE13 A0A194R6P4 A0A194QBL1 A0A212F4K9 Q8IRI6-10 Q8IRI6-2 A0A1L8DII3 T1P8T0 Q8IRI6-3 A0A2M4AR56 Q8IRI6-11 X2JBV9 W8AU68 A0A2M3Z9E3 W5JAR8 A0A2M4CJA9 A0A023EU33 A0A0S0WFC8 Q8IRI6-17 V9I6H3 A0A1Q3FWN3 A0A2A3EL61 Q17EP5 A0A2C9GPM1 A0A1L8DIT4 A0A1A9WPU6 X2JG36 Q8IRI6-16 Q7QJ79 Q8IRI6-12 A0A0J7KM10 Q8IRI6-14 Q8IRI6-6 Q8IRI6-15 A0A3L8DNS7 A0A026W3E5 A0A3B0JWX8 A0A1W4VUA3 A0A1Q3FX51 A0A2M4CUX3 B4IXB5 A0A1B6CWR9 B4QLH3 A0A087ZQD7 A0A1B6CTI4 A0A0L7R9B5 Q7PF87 A0A1I8MN39 A0A1I8PSE7 W0FVJ1 A0A1B6CQV5 A0A1B6DAY7 Q8IRI6-13 A0A023F5Q6 A0A1Z1XG22 T1IE15 B4HVL7 A0A0N8BMS5 A0A154PKQ2 A0A0N8EAH6 A0A146KMD7 A0A158NNR4 F4WV30 A0A164SYQ9 A0A0V0G868 A0A224XP36 A0A1B6CQC8 A0A1B6DC10 A0A069DV87 E9J9N5 A0A1B6EFA6 A0A1B6EAN5 B4KWH4 A0A146LVD3 A0A0Q9WIL2 A0A195BS38 A0A1B6DH78 A0A0P4VMS5 A0A1B6FH55 A0A1B6C2B8 A0A0K2UG24 A0A195EX69 A0A151WSZ8 A0A0P5C463 A0A2R5L7U2 A0A1B6LRT4 A0A0A8J8K9 A0A1B6L7Q0 A0A1B6J8Y5 A0A1B6MM35 A0A151IRI1 A0A0P5C448 A0A0P5C415 E2C2M2
Pubmed
19121390
28756777
26227816
26354079
22118469
10479996
+ More
10731132 12537572 12537569 12537568 12537573 12537574 16110336 17569856 17569867 24495485 20920257 23761445 24945155 17510324 12364791 14747013 17210077 30249741 24508170 17994087 25315136 24391959 25474469 28588501 26823975 21347285 21719571 26334808 21282665 27129103 20798317
10731132 12537572 12537569 12537568 12537573 12537574 16110336 17569856 17569867 24495485 20920257 23761445 24945155 17510324 12364791 14747013 17210077 30249741 24508170 17994087 25315136 24391959 25474469 28588501 26823975 21347285 21719571 26334808 21282665 27129103 20798317
EMBL
BABH01026028
KZ149893
PZC78941.1
NWSH01000685
PCG74784.1
PCG74785.1
+ More
PCG74786.1 ODYU01010734 SOQ56033.1 JTDY01001482 KOB73768.1 KQ460647 KPJ13189.1 KQ459249 KPJ02380.1 AGBW02010349 OWR48659.1 AF064703 AE014296 AY059441 BT011383 AAC36683.2 AAL13347.1 GFDF01007795 JAV06289.1 KA644535 AFP59164.1 GGFK01009940 MBW43261.1 AHN57921.1 GAMC01016783 GAMC01016782 JAB89772.1 GGFM01004405 MBW25156.1 ADMH02001654 ETN61547.1 GGFL01001239 MBW65417.1 GAPW01000720 JAC12878.1 ALI30448.1 JR035629 JR035630 AEY56555.1 AEY56556.1 GFDL01003014 JAV32031.1 KZ288223 PBC32012.1 CH477280 EAT44948.1 GFDF01007799 JAV06285.1 AHN57920.1 AAAB01008807 EAA04274.4 LBMM01005725 KMQ91266.1 QOIP01000006 RLU21458.1 KK107485 EZA50096.1 OUUW01000009 SPP84932.1 GFDL01003013 JAV32032.1 GGFL01004974 MBW69152.1 CH916366 EDV97447.1 GEDC01019533 JAS17765.1 CM000363 EDX08746.1 GEDC01020449 JAS16849.1 KQ414627 KOC67429.1 EAA45418.4 KF803268 AHF27422.1 GEDC01021510 JAS15788.1 GEDC01014486 JAS22812.1 GBBI01002207 JAC16505.1 KY350171 ARX98210.1 ACPB03000235 CH480817 EDW49982.1 GDIQ01162459 JAK89266.1 KQ434938 KZC12054.1 GDIQ01045860 JAN48877.1 GDHC01020905 JAP97723.1 ADTU01021729 ADTU01021730 ADTU01021731 ADTU01021732 GL888384 EGI61918.1 LRGB01001899 KZS10076.1 GECL01002228 JAP03896.1 GFTR01006667 JAW09759.1 GEDC01021687 JAS15611.1 GEDC01014065 JAS23233.1 GBGD01001272 JAC87617.1 GL769399 EFZ10419.1 GEDC01000676 JAS36622.1 GEDC01002354 JAS34944.1 CH933809 EDW19603.2 GDHC01007983 JAQ10646.1 CH940647 KRF84716.1 KQ976417 KYM89830.1 GEDC01012296 JAS25002.1 GDKW01002826 JAI53769.1 GECZ01020223 JAS49546.1 GEDC01029784 JAS07514.1 HACA01019526 CDW36887.1 KQ981953 KYN32497.1 KQ982766 KYQ50974.1 GDIP01175871 JAJ47531.1 GGLE01001423 MBY05549.1 GEBQ01013546 JAT26431.1 AB901049 BAQ02349.1 GEBQ01020250 JAT19727.1 GECU01012117 JAS95589.1 GEBQ01003006 JAT36971.1 KQ981124 KYN09323.1 GDIP01175870 JAJ47532.1 GDIP01175869 JAJ47533.1 GL452187 EFN77800.1
PCG74786.1 ODYU01010734 SOQ56033.1 JTDY01001482 KOB73768.1 KQ460647 KPJ13189.1 KQ459249 KPJ02380.1 AGBW02010349 OWR48659.1 AF064703 AE014296 AY059441 BT011383 AAC36683.2 AAL13347.1 GFDF01007795 JAV06289.1 KA644535 AFP59164.1 GGFK01009940 MBW43261.1 AHN57921.1 GAMC01016783 GAMC01016782 JAB89772.1 GGFM01004405 MBW25156.1 ADMH02001654 ETN61547.1 GGFL01001239 MBW65417.1 GAPW01000720 JAC12878.1 ALI30448.1 JR035629 JR035630 AEY56555.1 AEY56556.1 GFDL01003014 JAV32031.1 KZ288223 PBC32012.1 CH477280 EAT44948.1 GFDF01007799 JAV06285.1 AHN57920.1 AAAB01008807 EAA04274.4 LBMM01005725 KMQ91266.1 QOIP01000006 RLU21458.1 KK107485 EZA50096.1 OUUW01000009 SPP84932.1 GFDL01003013 JAV32032.1 GGFL01004974 MBW69152.1 CH916366 EDV97447.1 GEDC01019533 JAS17765.1 CM000363 EDX08746.1 GEDC01020449 JAS16849.1 KQ414627 KOC67429.1 EAA45418.4 KF803268 AHF27422.1 GEDC01021510 JAS15788.1 GEDC01014486 JAS22812.1 GBBI01002207 JAC16505.1 KY350171 ARX98210.1 ACPB03000235 CH480817 EDW49982.1 GDIQ01162459 JAK89266.1 KQ434938 KZC12054.1 GDIQ01045860 JAN48877.1 GDHC01020905 JAP97723.1 ADTU01021729 ADTU01021730 ADTU01021731 ADTU01021732 GL888384 EGI61918.1 LRGB01001899 KZS10076.1 GECL01002228 JAP03896.1 GFTR01006667 JAW09759.1 GEDC01021687 JAS15611.1 GEDC01014065 JAS23233.1 GBGD01001272 JAC87617.1 GL769399 EFZ10419.1 GEDC01000676 JAS36622.1 GEDC01002354 JAS34944.1 CH933809 EDW19603.2 GDHC01007983 JAQ10646.1 CH940647 KRF84716.1 KQ976417 KYM89830.1 GEDC01012296 JAS25002.1 GDKW01002826 JAI53769.1 GECZ01020223 JAS49546.1 GEDC01029784 JAS07514.1 HACA01019526 CDW36887.1 KQ981953 KYN32497.1 KQ982766 KYQ50974.1 GDIP01175871 JAJ47531.1 GGLE01001423 MBY05549.1 GEBQ01013546 JAT26431.1 AB901049 BAQ02349.1 GEBQ01020250 JAT19727.1 GECU01012117 JAS95589.1 GEBQ01003006 JAT36971.1 KQ981124 KYN09323.1 GDIP01175870 JAJ47532.1 GDIP01175869 JAJ47533.1 GL452187 EFN77800.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000053240
UP000053268
UP000007151
+ More
UP000000803 UP000000673 UP000242457 UP000008820 UP000091820 UP000007062 UP000036403 UP000279307 UP000053097 UP000268350 UP000192221 UP000001070 UP000000304 UP000005203 UP000053825 UP000095301 UP000095300 UP000015103 UP000001292 UP000076502 UP000005205 UP000007755 UP000076858 UP000009192 UP000008792 UP000078540 UP000078541 UP000075809 UP000078492 UP000008237
UP000000803 UP000000673 UP000242457 UP000008820 UP000091820 UP000007062 UP000036403 UP000279307 UP000053097 UP000268350 UP000192221 UP000001070 UP000000304 UP000005203 UP000053825 UP000095301 UP000095300 UP000015103 UP000001292 UP000076502 UP000005205 UP000007755 UP000076858 UP000009192 UP000008792 UP000078540 UP000078541 UP000075809 UP000078492 UP000008237
Pfam
PF00083 Sugar_tr
Interpro
SUPFAM
SSF103473
SSF103473
CDD
ProteinModelPortal
H9J9T0
A0A2W1BX91
A0A2A4JT42
A0A2A4JRX3
A0A2A4JSN4
A0A2H1WSK2
+ More
A0A0L7LE13 A0A194R6P4 A0A194QBL1 A0A212F4K9 Q8IRI6-10 Q8IRI6-2 A0A1L8DII3 T1P8T0 Q8IRI6-3 A0A2M4AR56 Q8IRI6-11 X2JBV9 W8AU68 A0A2M3Z9E3 W5JAR8 A0A2M4CJA9 A0A023EU33 A0A0S0WFC8 Q8IRI6-17 V9I6H3 A0A1Q3FWN3 A0A2A3EL61 Q17EP5 A0A2C9GPM1 A0A1L8DIT4 A0A1A9WPU6 X2JG36 Q8IRI6-16 Q7QJ79 Q8IRI6-12 A0A0J7KM10 Q8IRI6-14 Q8IRI6-6 Q8IRI6-15 A0A3L8DNS7 A0A026W3E5 A0A3B0JWX8 A0A1W4VUA3 A0A1Q3FX51 A0A2M4CUX3 B4IXB5 A0A1B6CWR9 B4QLH3 A0A087ZQD7 A0A1B6CTI4 A0A0L7R9B5 Q7PF87 A0A1I8MN39 A0A1I8PSE7 W0FVJ1 A0A1B6CQV5 A0A1B6DAY7 Q8IRI6-13 A0A023F5Q6 A0A1Z1XG22 T1IE15 B4HVL7 A0A0N8BMS5 A0A154PKQ2 A0A0N8EAH6 A0A146KMD7 A0A158NNR4 F4WV30 A0A164SYQ9 A0A0V0G868 A0A224XP36 A0A1B6CQC8 A0A1B6DC10 A0A069DV87 E9J9N5 A0A1B6EFA6 A0A1B6EAN5 B4KWH4 A0A146LVD3 A0A0Q9WIL2 A0A195BS38 A0A1B6DH78 A0A0P4VMS5 A0A1B6FH55 A0A1B6C2B8 A0A0K2UG24 A0A195EX69 A0A151WSZ8 A0A0P5C463 A0A2R5L7U2 A0A1B6LRT4 A0A0A8J8K9 A0A1B6L7Q0 A0A1B6J8Y5 A0A1B6MM35 A0A151IRI1 A0A0P5C448 A0A0P5C415 E2C2M2
A0A0L7LE13 A0A194R6P4 A0A194QBL1 A0A212F4K9 Q8IRI6-10 Q8IRI6-2 A0A1L8DII3 T1P8T0 Q8IRI6-3 A0A2M4AR56 Q8IRI6-11 X2JBV9 W8AU68 A0A2M3Z9E3 W5JAR8 A0A2M4CJA9 A0A023EU33 A0A0S0WFC8 Q8IRI6-17 V9I6H3 A0A1Q3FWN3 A0A2A3EL61 Q17EP5 A0A2C9GPM1 A0A1L8DIT4 A0A1A9WPU6 X2JG36 Q8IRI6-16 Q7QJ79 Q8IRI6-12 A0A0J7KM10 Q8IRI6-14 Q8IRI6-6 Q8IRI6-15 A0A3L8DNS7 A0A026W3E5 A0A3B0JWX8 A0A1W4VUA3 A0A1Q3FX51 A0A2M4CUX3 B4IXB5 A0A1B6CWR9 B4QLH3 A0A087ZQD7 A0A1B6CTI4 A0A0L7R9B5 Q7PF87 A0A1I8MN39 A0A1I8PSE7 W0FVJ1 A0A1B6CQV5 A0A1B6DAY7 Q8IRI6-13 A0A023F5Q6 A0A1Z1XG22 T1IE15 B4HVL7 A0A0N8BMS5 A0A154PKQ2 A0A0N8EAH6 A0A146KMD7 A0A158NNR4 F4WV30 A0A164SYQ9 A0A0V0G868 A0A224XP36 A0A1B6CQC8 A0A1B6DC10 A0A069DV87 E9J9N5 A0A1B6EFA6 A0A1B6EAN5 B4KWH4 A0A146LVD3 A0A0Q9WIL2 A0A195BS38 A0A1B6DH78 A0A0P4VMS5 A0A1B6FH55 A0A1B6C2B8 A0A0K2UG24 A0A195EX69 A0A151WSZ8 A0A0P5C463 A0A2R5L7U2 A0A1B6LRT4 A0A0A8J8K9 A0A1B6L7Q0 A0A1B6J8Y5 A0A1B6MM35 A0A151IRI1 A0A0P5C448 A0A0P5C415 E2C2M2
PDB
5EQI
E-value=5.25229e-105,
Score=974
Ontologies
GO
Topology
Subcellular location
Membrane
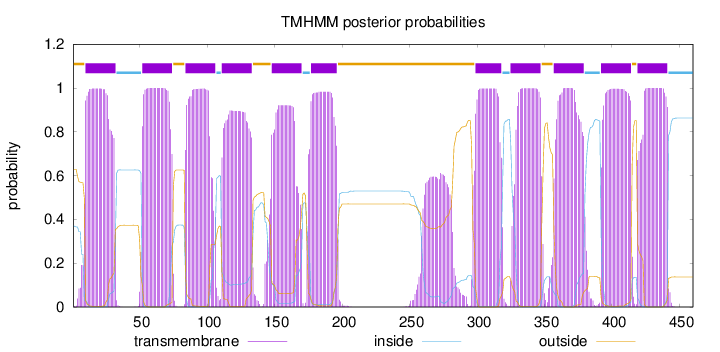
Length:
460
Number of predicted TMHs:
11
Exp number of AAs in TMHs:
250.57739
Exp number, first 60 AAs:
31.38045
Total prob of N-in:
0.37146
POSSIBLE N-term signal
sequence
outside
1 - 9
TMhelix
10 - 32
inside
33 - 51
TMhelix
52 - 74
outside
75 - 83
TMhelix
84 - 106
inside
107 - 110
TMhelix
111 - 133
outside
134 - 147
TMhelix
148 - 170
inside
171 - 176
TMhelix
177 - 196
outside
197 - 298
TMhelix
299 - 318
inside
319 - 324
TMhelix
325 - 347
outside
348 - 356
TMhelix
357 - 379
inside
380 - 391
TMhelix
392 - 414
outside
415 - 418
TMhelix
419 - 441
inside
442 - 460
Population Genetic Test Statistics
Pi
29.727212
Theta
25.854521
Tajima's D
0.269047
CLR
0.241769
CSRT
0.454777261136943
Interpretation
Uncertain
