Gene
KWMTBOMO03605
Pre Gene Modal
BGIBMGA006322
Annotation
PREDICTED:_soluble_guanylate_cyclase_89Db-like_[Bombyx_mori]
Full name
Guanylate cyclase
+ More
Soluble guanylate cyclase 89Db
Soluble guanylate cyclase 89Db
Location in the cell
PlasmaMembrane Reliability : 1.076
Sequence
CDS
ATGTACGGTTGGCTGTTGGAAAGCGTACAACATTTCATTGTGGTGGAGACAGTAGGAACAGTGTACATGCTGGTGTCCGGGGCGCCCGAGCGGCGCCGGGCTCATGCCGCTTCTGCCGCCAGTGCCGCGCTCGCAGTCTGCCGCGCCCTGCCCGCGCTTACTATAGGAATACATTCCGGTCCGTGCGTCGCGGGAGTGTTGGGGTTGAGGCTTCCTCGTTACTGTCTTGTGGGAGACACCGTGAACACAGCGAGTAGAATGCAAACTTCTTCTGAGCCGGGGCGGATCCAGATCTCCGCTACCACAGCGGCACAACTGCCCGCGGGAAGGTTCAGGCTTCGTCCTCGAGGTGTTATCAAAGTTAAGGGCAAGGGCATGATGGAAACGTTCTGGTTGGAAGGAGAGATCGAAGAAGAGGAACAAGACGAGGCCTTGCAGCTGTTCTCGGCTTTATGCGGAGACGATTAA
Protein
MYGWLLESVQHFIVVETVGTVYMLVSGAPERRRAHAASAASAALAVCRALPALTIGIHSGPCVAGVLGLRLPRYCLVGDTVNTASRMQTSSEPGRIQISATTAAQLPAGRFRLRPRGVIKVKGKGMMETFWLEGEIEEEEQDEALQLFSALCGDD
Summary
Description
Heterodimers with Gyc88E are activated in response to changing oxygen concentrations, alerting flies to hypoxic environments. Under normal oxygen concentrations, oxygen binds to the heme group and results in low levels of guanylyl cyclase activity. When exposed to reduced oxygen concentrations, the oxygen dissociates from the heme group resulting in activation of the enzyme.
Catalytic Activity
GTP = 3',5'-cyclic GMP + diphosphate
Cofactor
heme
Subunit
Heterodimer; with Gyc88E, in the presence of magnesium or manganese.
Miscellaneous
There are two types of guanylate cyclases: soluble forms and membrane-associated receptor forms.
Similarity
Belongs to the adenylyl cyclase class-4/guanylyl cyclase family.
Belongs to the peptidase S1 family.
Belongs to the peptidase S1 family.
Keywords
cGMP biosynthesis
Coiled coil
Complete proteome
Cytoplasm
GTP-binding
Heme
Iron
Lyase
Magnesium
Manganese
Metal-binding
Nucleotide-binding
Reference proteome
Feature
chain Guanylate cyclase
Uniprot
A0A2H1WEH5
A0A2W1BV30
A0A212F4L0
A0A2A4JT61
A0A194R7H4
F4X0S1
+ More
A0A195EIX4 A0A195D4S4 A0A195F4F4 T1GJF6 E2BDU0 A0A2S2P608 A0A158P3G7 A0A087UVT7 A0A2J7Q455 A0A154PE10 A0A0C9PMU3 E2AD43 A0A0L7QZC3 J9K961 A0A2J7Q469 A0A2J7Q456 A0A1Y1KDT5 K7J299 D6W9P2 A0A1Y1KBV4 A0A1A9X1X7 A0A0K8U9F2 A0A1B0C4E3 A0A034WN86 T1G4Y1 U4UVZ5 A0A0C9QG99 A0A1W0WK94 A0A1B0D034 A0A2A3E5P7 A0A087ZRI8 A0A1B0F9W4 A0A1A9VS86 A0A0M4F2H5 B4NK75 B3M2T1 A0A210QXS7 A0A0A1XJC8 A0A091H8N2 A0A093J077 A0A182GRH5 A0A1I8PFH4 A0A093FJC9 A0A0L0BPN4 A0A3B0JT95 W8BWP3 B4JFC2 H9GJY6 B4GFC0 Q296C7 A0A093NT13 A0A087QL28 A0A0N4VCG2 A0A194RMG7 B4IBU8 R7TGC9 A0A1W4U765 B4QX22 Q9VEU5 A0A194PLI8 B4K682 A0A336LM52 A0A131ZXH9 A0A212FC80 A0A091SFK1 A0A1I8MS99 A0A182F8Z5 A0A336KIL9 W4YIH3 Q16KE1 A0A1Y1MV38 A0A182J8R8
A0A195EIX4 A0A195D4S4 A0A195F4F4 T1GJF6 E2BDU0 A0A2S2P608 A0A158P3G7 A0A087UVT7 A0A2J7Q455 A0A154PE10 A0A0C9PMU3 E2AD43 A0A0L7QZC3 J9K961 A0A2J7Q469 A0A2J7Q456 A0A1Y1KDT5 K7J299 D6W9P2 A0A1Y1KBV4 A0A1A9X1X7 A0A0K8U9F2 A0A1B0C4E3 A0A034WN86 T1G4Y1 U4UVZ5 A0A0C9QG99 A0A1W0WK94 A0A1B0D034 A0A2A3E5P7 A0A087ZRI8 A0A1B0F9W4 A0A1A9VS86 A0A0M4F2H5 B4NK75 B3M2T1 A0A210QXS7 A0A0A1XJC8 A0A091H8N2 A0A093J077 A0A182GRH5 A0A1I8PFH4 A0A093FJC9 A0A0L0BPN4 A0A3B0JT95 W8BWP3 B4JFC2 H9GJY6 B4GFC0 Q296C7 A0A093NT13 A0A087QL28 A0A0N4VCG2 A0A194RMG7 B4IBU8 R7TGC9 A0A1W4U765 B4QX22 Q9VEU5 A0A194PLI8 B4K682 A0A336LM52 A0A131ZXH9 A0A212FC80 A0A091SFK1 A0A1I8MS99 A0A182F8Z5 A0A336KIL9 W4YIH3 Q16KE1 A0A1Y1MV38 A0A182J8R8
EC Number
4.6.1.2
Pubmed
EMBL
ODYU01007806
SOQ50884.1
KZ149893
PZC78942.1
AGBW02010349
OWR48660.1
+ More
NWSH01000685 PCG74804.1 KQ460647 KPJ13190.1 GL888499 EGI59948.1 KQ978801 KYN28210.1 KQ976842 KYN07905.1 KQ981805 KYN35460.1 CAQQ02139368 CAQQ02139369 CAQQ02139370 GL447689 EFN86146.1 GGMR01012280 MBY24899.1 ADTU01007948 ADTU01007949 ADTU01007950 KK121891 KFM81476.1 NEVH01018390 PNF23371.1 KQ434869 KZC09430.1 GBYB01002468 JAG72235.1 GL438663 EFN68637.1 KQ414681 KOC63970.1 ABLF02030871 ABLF02030879 PNF23370.1 PNF23372.1 GEZM01091311 JAV56927.1 AAZX01006360 KQ971312 EEZ98124.2 GEZM01091312 JAV56926.1 GDHF01029324 JAI22990.1 JXJN01025424 GAKP01003327 JAC55625.1 AMQM01005395 KB096900 ESO00696.1 KB632390 ERL94426.1 GBYB01002469 JAG72236.1 MTYJ01000087 OQV15543.1 AJVK01009807 KZ288360 PBC27018.1 CCAG010010254 CP012526 ALC45282.1 CH964272 EDW85117.1 CH902617 EDV43461.1 NEDP02001335 OWF53513.1 GBXI01003664 JAD10628.1 KL525367 KFO90995.1 KK566140 KFW04414.1 JXUM01082520 KQ563318 KXJ74066.1 KK390609 KFV54421.1 JRES01001565 KNC22017.1 OUUW01000007 SPP83622.1 GAMC01012927 GAMC01012926 JAB93629.1 CH916369 EDV93403.1 CH479182 EDW34305.1 CM000070 EAL28531.1 KRT00794.1 KL225141 KFW67948.1 KL225737 KFM01932.1 UXUI01009068 VDD92993.1 KQ459995 KPJ18525.1 CH480827 EDW44856.1 AMQN01013154 AMQN01013155 KB309995 ELT92823.1 CM000364 EDX12676.1 AE014297 AY051421 AAF55323.1 AAK92845.1 KQ459600 KPI94301.1 CH933806 EDW14132.1 KRG00873.1 UFQT01000054 SSX19066.1 JXLN01005296 KPM03516.1 AGBW02009215 OWR51328.1 KK470231 KFQ57168.1 UFQS01000473 UFQT01000473 SSX04267.1 SSX24632.1 AAGJ04150183 AAGJ04150184 CH477963 EAT34765.1 GEZM01019893 JAV89543.1
NWSH01000685 PCG74804.1 KQ460647 KPJ13190.1 GL888499 EGI59948.1 KQ978801 KYN28210.1 KQ976842 KYN07905.1 KQ981805 KYN35460.1 CAQQ02139368 CAQQ02139369 CAQQ02139370 GL447689 EFN86146.1 GGMR01012280 MBY24899.1 ADTU01007948 ADTU01007949 ADTU01007950 KK121891 KFM81476.1 NEVH01018390 PNF23371.1 KQ434869 KZC09430.1 GBYB01002468 JAG72235.1 GL438663 EFN68637.1 KQ414681 KOC63970.1 ABLF02030871 ABLF02030879 PNF23370.1 PNF23372.1 GEZM01091311 JAV56927.1 AAZX01006360 KQ971312 EEZ98124.2 GEZM01091312 JAV56926.1 GDHF01029324 JAI22990.1 JXJN01025424 GAKP01003327 JAC55625.1 AMQM01005395 KB096900 ESO00696.1 KB632390 ERL94426.1 GBYB01002469 JAG72236.1 MTYJ01000087 OQV15543.1 AJVK01009807 KZ288360 PBC27018.1 CCAG010010254 CP012526 ALC45282.1 CH964272 EDW85117.1 CH902617 EDV43461.1 NEDP02001335 OWF53513.1 GBXI01003664 JAD10628.1 KL525367 KFO90995.1 KK566140 KFW04414.1 JXUM01082520 KQ563318 KXJ74066.1 KK390609 KFV54421.1 JRES01001565 KNC22017.1 OUUW01000007 SPP83622.1 GAMC01012927 GAMC01012926 JAB93629.1 CH916369 EDV93403.1 CH479182 EDW34305.1 CM000070 EAL28531.1 KRT00794.1 KL225141 KFW67948.1 KL225737 KFM01932.1 UXUI01009068 VDD92993.1 KQ459995 KPJ18525.1 CH480827 EDW44856.1 AMQN01013154 AMQN01013155 KB309995 ELT92823.1 CM000364 EDX12676.1 AE014297 AY051421 AAF55323.1 AAK92845.1 KQ459600 KPI94301.1 CH933806 EDW14132.1 KRG00873.1 UFQT01000054 SSX19066.1 JXLN01005296 KPM03516.1 AGBW02009215 OWR51328.1 KK470231 KFQ57168.1 UFQS01000473 UFQT01000473 SSX04267.1 SSX24632.1 AAGJ04150183 AAGJ04150184 CH477963 EAT34765.1 GEZM01019893 JAV89543.1
Proteomes
UP000007151
UP000218220
UP000053240
UP000007755
UP000078492
UP000078542
+ More
UP000078541 UP000015102 UP000008237 UP000005205 UP000054359 UP000235965 UP000076502 UP000000311 UP000053825 UP000007819 UP000002358 UP000007266 UP000091820 UP000092460 UP000015101 UP000030742 UP000092462 UP000242457 UP000005203 UP000092444 UP000078200 UP000092553 UP000007798 UP000007801 UP000242188 UP000069940 UP000249989 UP000095300 UP000037069 UP000268350 UP000001070 UP000001646 UP000008744 UP000001819 UP000054081 UP000053286 UP000038041 UP000274131 UP000001292 UP000014760 UP000192221 UP000000304 UP000000803 UP000053268 UP000009192 UP000095301 UP000069272 UP000007110 UP000008820 UP000075880
UP000078541 UP000015102 UP000008237 UP000005205 UP000054359 UP000235965 UP000076502 UP000000311 UP000053825 UP000007819 UP000002358 UP000007266 UP000091820 UP000092460 UP000015101 UP000030742 UP000092462 UP000242457 UP000005203 UP000092444 UP000078200 UP000092553 UP000007798 UP000007801 UP000242188 UP000069940 UP000249989 UP000095300 UP000037069 UP000268350 UP000001070 UP000001646 UP000008744 UP000001819 UP000054081 UP000053286 UP000038041 UP000274131 UP000001292 UP000014760 UP000192221 UP000000304 UP000000803 UP000053268 UP000009192 UP000095301 UP000069272 UP000007110 UP000008820 UP000075880
PRIDE
Pfam
Interpro
IPR029787
Nucleotide_cyclase
+ More
IPR011645 HNOB_dom_associated
IPR001054 A/G_cyclase
IPR038158 H-NOX_domain_sf
IPR024096 NO_sig/Golgi_transp_ligand-bd
IPR011644 Heme_NO-bd
IPR018297 A/G_cyclase_CS
IPR009003 Peptidase_S1_PA
IPR001314 Peptidase_S1A
IPR001254 Trypsin_dom
IPR018114 TRYPSIN_HIS
IPR033116 TRYPSIN_SER
IPR011009 Kinase-like_dom_sf
IPR001245 Ser-Thr/Tyr_kinase_cat_dom
IPR001828 ANF_lig-bd_rcpt
IPR000719 Prot_kinase_dom
IPR028082 Peripla_BP_I
IPR001170 ANPR/GUC
IPR013587 Nitrate/nitrite_sensing
IPR011645 HNOB_dom_associated
IPR001054 A/G_cyclase
IPR038158 H-NOX_domain_sf
IPR024096 NO_sig/Golgi_transp_ligand-bd
IPR011644 Heme_NO-bd
IPR018297 A/G_cyclase_CS
IPR009003 Peptidase_S1_PA
IPR001314 Peptidase_S1A
IPR001254 Trypsin_dom
IPR018114 TRYPSIN_HIS
IPR033116 TRYPSIN_SER
IPR011009 Kinase-like_dom_sf
IPR001245 Ser-Thr/Tyr_kinase_cat_dom
IPR001828 ANF_lig-bd_rcpt
IPR000719 Prot_kinase_dom
IPR028082 Peripla_BP_I
IPR001170 ANPR/GUC
IPR013587 Nitrate/nitrite_sensing
SUPFAM
Gene 3D
CDD
ProteinModelPortal
A0A2H1WEH5
A0A2W1BV30
A0A212F4L0
A0A2A4JT61
A0A194R7H4
F4X0S1
+ More
A0A195EIX4 A0A195D4S4 A0A195F4F4 T1GJF6 E2BDU0 A0A2S2P608 A0A158P3G7 A0A087UVT7 A0A2J7Q455 A0A154PE10 A0A0C9PMU3 E2AD43 A0A0L7QZC3 J9K961 A0A2J7Q469 A0A2J7Q456 A0A1Y1KDT5 K7J299 D6W9P2 A0A1Y1KBV4 A0A1A9X1X7 A0A0K8U9F2 A0A1B0C4E3 A0A034WN86 T1G4Y1 U4UVZ5 A0A0C9QG99 A0A1W0WK94 A0A1B0D034 A0A2A3E5P7 A0A087ZRI8 A0A1B0F9W4 A0A1A9VS86 A0A0M4F2H5 B4NK75 B3M2T1 A0A210QXS7 A0A0A1XJC8 A0A091H8N2 A0A093J077 A0A182GRH5 A0A1I8PFH4 A0A093FJC9 A0A0L0BPN4 A0A3B0JT95 W8BWP3 B4JFC2 H9GJY6 B4GFC0 Q296C7 A0A093NT13 A0A087QL28 A0A0N4VCG2 A0A194RMG7 B4IBU8 R7TGC9 A0A1W4U765 B4QX22 Q9VEU5 A0A194PLI8 B4K682 A0A336LM52 A0A131ZXH9 A0A212FC80 A0A091SFK1 A0A1I8MS99 A0A182F8Z5 A0A336KIL9 W4YIH3 Q16KE1 A0A1Y1MV38 A0A182J8R8
A0A195EIX4 A0A195D4S4 A0A195F4F4 T1GJF6 E2BDU0 A0A2S2P608 A0A158P3G7 A0A087UVT7 A0A2J7Q455 A0A154PE10 A0A0C9PMU3 E2AD43 A0A0L7QZC3 J9K961 A0A2J7Q469 A0A2J7Q456 A0A1Y1KDT5 K7J299 D6W9P2 A0A1Y1KBV4 A0A1A9X1X7 A0A0K8U9F2 A0A1B0C4E3 A0A034WN86 T1G4Y1 U4UVZ5 A0A0C9QG99 A0A1W0WK94 A0A1B0D034 A0A2A3E5P7 A0A087ZRI8 A0A1B0F9W4 A0A1A9VS86 A0A0M4F2H5 B4NK75 B3M2T1 A0A210QXS7 A0A0A1XJC8 A0A091H8N2 A0A093J077 A0A182GRH5 A0A1I8PFH4 A0A093FJC9 A0A0L0BPN4 A0A3B0JT95 W8BWP3 B4JFC2 H9GJY6 B4GFC0 Q296C7 A0A093NT13 A0A087QL28 A0A0N4VCG2 A0A194RMG7 B4IBU8 R7TGC9 A0A1W4U765 B4QX22 Q9VEU5 A0A194PLI8 B4K682 A0A336LM52 A0A131ZXH9 A0A212FC80 A0A091SFK1 A0A1I8MS99 A0A182F8Z5 A0A336KIL9 W4YIH3 Q16KE1 A0A1Y1MV38 A0A182J8R8
PDB
3ET6
E-value=1.89485e-17,
Score=213
Ontologies
PATHWAY
GO
GO:0004383
GO:0035556
GO:0020037
GO:0016021
GO:0000166
GO:0016849
GO:0009190
GO:0004252
GO:0007165
GO:0006182
GO:0007168
GO:0005886
GO:0001653
GO:0005524
GO:0004672
GO:0001666
GO:0038060
GO:0046982
GO:0008074
GO:0000302
GO:0055093
GO:0005525
GO:0019934
GO:0046872
GO:0051103
GO:0008270
GO:0042555
GO:0048384
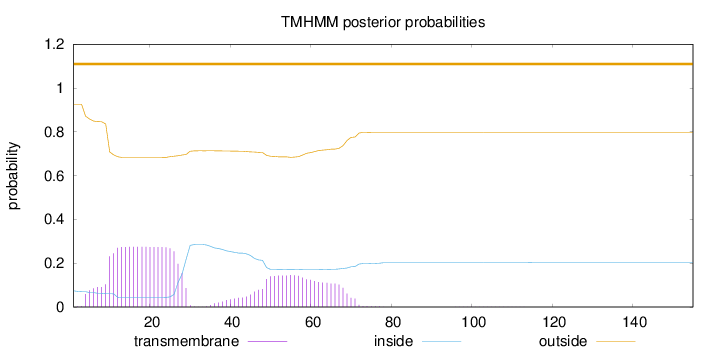
Topology
Subcellular location
Cytoplasm
Length:
155
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
8.7986
Exp number, first 60 AAs:
7.7704
Total prob of N-in:
0.07407
outside
1 - 155
Population Genetic Test Statistics
Pi
32.044701
Theta
29.238103
Tajima's D
0.045435
CLR
0.429104
CSRT
0.380930953452327
Interpretation
Uncertain
