Gene
KWMTBOMO03600 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA006318
Annotation
farnesoic_acid_O-methyltransferase_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.946
Sequence
CDS
ATGGCAGGCACATATCGTTGGGTTCCTGCTTGCTTAAATCAAGGCAGCATTCCACCAGGTGCCCTCAGAGTCGGTACCGATGCCGATGGAGACGAAATCTACGCTGGAAGAGCTCCTCACGAGGGTGAAATTGTTCCCGCCAAGGTTATTCCGAACAAGAACGTTTGCTTTATATCGTTCTGCGGCGAGGAAATTTTAAAGGACCAGTTTGAGGTGCTTGTTCCGACTTTCTTCGCTTGGCAGTTTGCCTCGAACGGTGAACTTCCCCCTGGTGCAGTGAACTTTGGTGTCACAGCCGATGGTGAGAGACTCTACTACGGTAGGGTCACTCGCGATGGACTGACGACGCCTGGCAAGATTCAACCTTCGCATGGTTGCTGTTATTTTCCCCTCGATGGCGGAGAACAGAGCTCCACTGAATATGAATGTTTGGTGCTTATATAA
Protein
MAGTYRWVPACLNQGSIPPGALRVGTDADGDEIYAGRAPHEGEIVPAKVIPNKNVCFISFCGEEILKDQFEVLVPTFFAWQFASNGELPPGAVNFGVTADGERLYYGRVTRDGLTTPGKIQPSHGCCYFPLDGGEQSSTEYECLVLI
Summary
Uniprot
S5SA62
H9J9X3
S5RVX1
S5RN30
A0A194QAC5
Q0PKS1
+ More
S5SBB2 A0A2H1VNW9 A0A212ENC0 A0A194R5U7 E3UKK4 A0A1E1W2N2 A0A0K8TFQ3 A0A212FL40 A0A2A4JSJ2 A0A2W1C0A5 A0A2A4JRY0 H9J9X4 A0A2A4JSX9 A0A0L7L642 A0A182Q766 U5EST7 A0A2M4CY56 A0A2M3Z0L1 A0A2M4CXN9 Q7Q607 T1DTN5 W5JNC4 A7UU14 A0A2M4A4N8 A0A182TWP0 A0A2M3Z0M1 A0A2M4A207 A0A182UM33 A0A182IKE0 A0A182WHU3 A0A182T8G4 A0A182NM71 A0A2M4A1N1 A0A2M4A1C2 A0A2M4C2L5 A0A2M4C2T8 A7UU13 A0A0J7KNB4 Q17AK6 A0A182RGT9 A0A182XP53 A0A182I1H2 A0A182K6L9 Q17AK5 A0A1Q3FP75 A0A2M4CHZ8 A0A2M4BZT2 A0A2M4BZD0 Q17AK7 A0A1S4FA09 B0WXL4 A0A182P102 A0A1S4FA24 A0A182M7T9 A0A182HDP5 A0A1B0CT69 A0A336MYF6 A0A182Y347 A0A0K8TQR4 A0A336M4D0 A0A336K3V4 E9IUT8 K7J2K3 A0A195AZ31 A0A151IT85 E2A658 A0A158NZ59 A0A1L8DCM8 A0A1L8DDL9 W5JCW9 A0A026WNN3 A0A2M4AY15 A0A182FHR1 A0A2M3Z8U3 A0A2M3Z9J5 A0A1J1IHI7 R4WCK1 A0A182FDR8 E2BSZ6 A0A182WC92 R4WCX6 A0A182NZ82 A0A151WHQ7 A0A1L8DCV1 U5EPK1 A0A182YLW5 A0A182M9U4 A0A2A3EBR4 A0A182R5I9 A8C9Z5 T1HAB7 A0A182QA83 A0A182PEU1 A0A0A9YX12
S5SBB2 A0A2H1VNW9 A0A212ENC0 A0A194R5U7 E3UKK4 A0A1E1W2N2 A0A0K8TFQ3 A0A212FL40 A0A2A4JSJ2 A0A2W1C0A5 A0A2A4JRY0 H9J9X4 A0A2A4JSX9 A0A0L7L642 A0A182Q766 U5EST7 A0A2M4CY56 A0A2M3Z0L1 A0A2M4CXN9 Q7Q607 T1DTN5 W5JNC4 A7UU14 A0A2M4A4N8 A0A182TWP0 A0A2M3Z0M1 A0A2M4A207 A0A182UM33 A0A182IKE0 A0A182WHU3 A0A182T8G4 A0A182NM71 A0A2M4A1N1 A0A2M4A1C2 A0A2M4C2L5 A0A2M4C2T8 A7UU13 A0A0J7KNB4 Q17AK6 A0A182RGT9 A0A182XP53 A0A182I1H2 A0A182K6L9 Q17AK5 A0A1Q3FP75 A0A2M4CHZ8 A0A2M4BZT2 A0A2M4BZD0 Q17AK7 A0A1S4FA09 B0WXL4 A0A182P102 A0A1S4FA24 A0A182M7T9 A0A182HDP5 A0A1B0CT69 A0A336MYF6 A0A182Y347 A0A0K8TQR4 A0A336M4D0 A0A336K3V4 E9IUT8 K7J2K3 A0A195AZ31 A0A151IT85 E2A658 A0A158NZ59 A0A1L8DCM8 A0A1L8DDL9 W5JCW9 A0A026WNN3 A0A2M4AY15 A0A182FHR1 A0A2M3Z8U3 A0A2M3Z9J5 A0A1J1IHI7 R4WCK1 A0A182FDR8 E2BSZ6 A0A182WC92 R4WCX6 A0A182NZ82 A0A151WHQ7 A0A1L8DCV1 U5EPK1 A0A182YLW5 A0A182M9U4 A0A2A3EBR4 A0A182R5I9 A8C9Z5 T1HAB7 A0A182QA83 A0A182PEU1 A0A0A9YX12
Pubmed
EMBL
KC888748
AGS17913.1
BABH01026017
KC888752
AGS17917.1
KC888749
+ More
AGS17914.1 KQ459249 KPJ02384.1 DQ813502 ABG67688.1 KC888750 AGS17915.1 ODYU01003568 SOQ42498.1 AGBW02013676 OWR42992.1 KQ460647 KPJ13193.1 HM449878 ADO33013.1 GDQN01009819 JAT81235.1 GBRD01001403 JAG64418.1 AGBW02007937 OWR54409.1 NWSH01000685 PCG74799.1 KZ149893 PZC78947.1 PCG74797.1 BABH01026018 PCG74798.1 JTDY01002686 KOB70922.1 AXCN02000945 GANO01004684 JAB55187.1 GGFL01006017 MBW70195.1 GGFM01001227 MBW21978.1 GGFL01005922 MBW70100.1 AAAB01008960 EAA11278.4 GAMD01000849 JAB00742.1 ADMH02001054 ETN64274.1 EDO63791.1 GGFK01002432 MBW35753.1 GGFM01001314 MBW22065.1 GGFK01001468 MBW34789.1 GGFK01001227 MBW34548.1 GGFK01001228 MBW34549.1 GGFJ01010426 MBW59567.1 GGFJ01010330 MBW59471.1 EDO63790.1 LBMM01005114 KMQ91802.1 CH477333 EAT43284.1 EAT43286.1 GFDL01005762 JAV29283.1 GGFL01000677 MBW64855.1 GGFJ01009373 MBW58514.1 GGFJ01009296 MBW58437.1 EAT43285.1 DS232168 EDS36544.1 AXCM01017129 JXUM01034614 JXUM01034615 JXUM01034616 KQ561030 KXJ79965.1 AJWK01027190 AJWK01027191 AJWK01027192 UFQS01003168 UFQT01003168 SSX15306.1 SSX34681.1 GDAI01001358 JAI16245.1 UFQT01000381 SSX23783.1 UFQS01000040 UFQT01000040 SSW98248.1 SSX18634.1 GL766050 EFZ15666.1 KQ976701 KYM77297.1 KQ981029 KYN10188.1 GL437108 EFN71117.1 ADTU01003883 GFDF01009866 JAV04218.1 GFDF01009609 JAV04475.1 ADMH02001746 ETN61198.1 KK107151 QOIP01000003 EZA57286.1 RLU25055.1 GGFK01012362 MBW45683.1 GGFM01004192 MBW24943.1 GGFM01004357 MBW25108.1 CVRI01000052 CRK99717.1 AK416938 BAN20153.1 GL450313 EFN81184.1 AK417024 BAN20239.1 KQ983114 KYQ47335.1 GFDF01009934 JAV04150.1 GANO01004728 JAB55143.1 AXCM01003759 KZ288292 PBC29177.1 EU035819 ABV44721.1 ACPB03019349 AXCN02000886 GBHO01007991 JAG35613.1
AGS17914.1 KQ459249 KPJ02384.1 DQ813502 ABG67688.1 KC888750 AGS17915.1 ODYU01003568 SOQ42498.1 AGBW02013676 OWR42992.1 KQ460647 KPJ13193.1 HM449878 ADO33013.1 GDQN01009819 JAT81235.1 GBRD01001403 JAG64418.1 AGBW02007937 OWR54409.1 NWSH01000685 PCG74799.1 KZ149893 PZC78947.1 PCG74797.1 BABH01026018 PCG74798.1 JTDY01002686 KOB70922.1 AXCN02000945 GANO01004684 JAB55187.1 GGFL01006017 MBW70195.1 GGFM01001227 MBW21978.1 GGFL01005922 MBW70100.1 AAAB01008960 EAA11278.4 GAMD01000849 JAB00742.1 ADMH02001054 ETN64274.1 EDO63791.1 GGFK01002432 MBW35753.1 GGFM01001314 MBW22065.1 GGFK01001468 MBW34789.1 GGFK01001227 MBW34548.1 GGFK01001228 MBW34549.1 GGFJ01010426 MBW59567.1 GGFJ01010330 MBW59471.1 EDO63790.1 LBMM01005114 KMQ91802.1 CH477333 EAT43284.1 EAT43286.1 GFDL01005762 JAV29283.1 GGFL01000677 MBW64855.1 GGFJ01009373 MBW58514.1 GGFJ01009296 MBW58437.1 EAT43285.1 DS232168 EDS36544.1 AXCM01017129 JXUM01034614 JXUM01034615 JXUM01034616 KQ561030 KXJ79965.1 AJWK01027190 AJWK01027191 AJWK01027192 UFQS01003168 UFQT01003168 SSX15306.1 SSX34681.1 GDAI01001358 JAI16245.1 UFQT01000381 SSX23783.1 UFQS01000040 UFQT01000040 SSW98248.1 SSX18634.1 GL766050 EFZ15666.1 KQ976701 KYM77297.1 KQ981029 KYN10188.1 GL437108 EFN71117.1 ADTU01003883 GFDF01009866 JAV04218.1 GFDF01009609 JAV04475.1 ADMH02001746 ETN61198.1 KK107151 QOIP01000003 EZA57286.1 RLU25055.1 GGFK01012362 MBW45683.1 GGFM01004192 MBW24943.1 GGFM01004357 MBW25108.1 CVRI01000052 CRK99717.1 AK416938 BAN20153.1 GL450313 EFN81184.1 AK417024 BAN20239.1 KQ983114 KYQ47335.1 GFDF01009934 JAV04150.1 GANO01004728 JAB55143.1 AXCM01003759 KZ288292 PBC29177.1 EU035819 ABV44721.1 ACPB03019349 AXCN02000886 GBHO01007991 JAG35613.1
Proteomes
UP000005204
UP000053268
UP000007151
UP000053240
UP000218220
UP000037510
+ More
UP000075886 UP000007062 UP000000673 UP000075902 UP000075903 UP000075880 UP000075920 UP000075901 UP000075884 UP000036403 UP000008820 UP000075900 UP000076407 UP000075881 UP000002320 UP000075885 UP000075883 UP000069940 UP000249989 UP000092461 UP000076408 UP000002358 UP000078540 UP000078492 UP000000311 UP000005205 UP000053097 UP000279307 UP000069272 UP000183832 UP000008237 UP000075809 UP000242457 UP000015103
UP000075886 UP000007062 UP000000673 UP000075902 UP000075903 UP000075880 UP000075920 UP000075901 UP000075884 UP000036403 UP000008820 UP000075900 UP000076407 UP000075881 UP000002320 UP000075885 UP000075883 UP000069940 UP000249989 UP000092461 UP000076408 UP000002358 UP000078540 UP000078492 UP000000311 UP000005205 UP000053097 UP000279307 UP000069272 UP000183832 UP000008237 UP000075809 UP000242457 UP000015103
Pfam
PF11901 DUF3421
Interpro
SUPFAM
SSF57424
SSF57424
Gene 3D
CDD
ProteinModelPortal
S5SA62
H9J9X3
S5RVX1
S5RN30
A0A194QAC5
Q0PKS1
+ More
S5SBB2 A0A2H1VNW9 A0A212ENC0 A0A194R5U7 E3UKK4 A0A1E1W2N2 A0A0K8TFQ3 A0A212FL40 A0A2A4JSJ2 A0A2W1C0A5 A0A2A4JRY0 H9J9X4 A0A2A4JSX9 A0A0L7L642 A0A182Q766 U5EST7 A0A2M4CY56 A0A2M3Z0L1 A0A2M4CXN9 Q7Q607 T1DTN5 W5JNC4 A7UU14 A0A2M4A4N8 A0A182TWP0 A0A2M3Z0M1 A0A2M4A207 A0A182UM33 A0A182IKE0 A0A182WHU3 A0A182T8G4 A0A182NM71 A0A2M4A1N1 A0A2M4A1C2 A0A2M4C2L5 A0A2M4C2T8 A7UU13 A0A0J7KNB4 Q17AK6 A0A182RGT9 A0A182XP53 A0A182I1H2 A0A182K6L9 Q17AK5 A0A1Q3FP75 A0A2M4CHZ8 A0A2M4BZT2 A0A2M4BZD0 Q17AK7 A0A1S4FA09 B0WXL4 A0A182P102 A0A1S4FA24 A0A182M7T9 A0A182HDP5 A0A1B0CT69 A0A336MYF6 A0A182Y347 A0A0K8TQR4 A0A336M4D0 A0A336K3V4 E9IUT8 K7J2K3 A0A195AZ31 A0A151IT85 E2A658 A0A158NZ59 A0A1L8DCM8 A0A1L8DDL9 W5JCW9 A0A026WNN3 A0A2M4AY15 A0A182FHR1 A0A2M3Z8U3 A0A2M3Z9J5 A0A1J1IHI7 R4WCK1 A0A182FDR8 E2BSZ6 A0A182WC92 R4WCX6 A0A182NZ82 A0A151WHQ7 A0A1L8DCV1 U5EPK1 A0A182YLW5 A0A182M9U4 A0A2A3EBR4 A0A182R5I9 A8C9Z5 T1HAB7 A0A182QA83 A0A182PEU1 A0A0A9YX12
S5SBB2 A0A2H1VNW9 A0A212ENC0 A0A194R5U7 E3UKK4 A0A1E1W2N2 A0A0K8TFQ3 A0A212FL40 A0A2A4JSJ2 A0A2W1C0A5 A0A2A4JRY0 H9J9X4 A0A2A4JSX9 A0A0L7L642 A0A182Q766 U5EST7 A0A2M4CY56 A0A2M3Z0L1 A0A2M4CXN9 Q7Q607 T1DTN5 W5JNC4 A7UU14 A0A2M4A4N8 A0A182TWP0 A0A2M3Z0M1 A0A2M4A207 A0A182UM33 A0A182IKE0 A0A182WHU3 A0A182T8G4 A0A182NM71 A0A2M4A1N1 A0A2M4A1C2 A0A2M4C2L5 A0A2M4C2T8 A7UU13 A0A0J7KNB4 Q17AK6 A0A182RGT9 A0A182XP53 A0A182I1H2 A0A182K6L9 Q17AK5 A0A1Q3FP75 A0A2M4CHZ8 A0A2M4BZT2 A0A2M4BZD0 Q17AK7 A0A1S4FA09 B0WXL4 A0A182P102 A0A1S4FA24 A0A182M7T9 A0A182HDP5 A0A1B0CT69 A0A336MYF6 A0A182Y347 A0A0K8TQR4 A0A336M4D0 A0A336K3V4 E9IUT8 K7J2K3 A0A195AZ31 A0A151IT85 E2A658 A0A158NZ59 A0A1L8DCM8 A0A1L8DDL9 W5JCW9 A0A026WNN3 A0A2M4AY15 A0A182FHR1 A0A2M3Z8U3 A0A2M3Z9J5 A0A1J1IHI7 R4WCK1 A0A182FDR8 E2BSZ6 A0A182WC92 R4WCX6 A0A182NZ82 A0A151WHQ7 A0A1L8DCV1 U5EPK1 A0A182YLW5 A0A182M9U4 A0A2A3EBR4 A0A182R5I9 A8C9Z5 T1HAB7 A0A182QA83 A0A182PEU1 A0A0A9YX12
PDB
5MH1
E-value=2.62184e-13,
Score=176
Ontologies
Topology
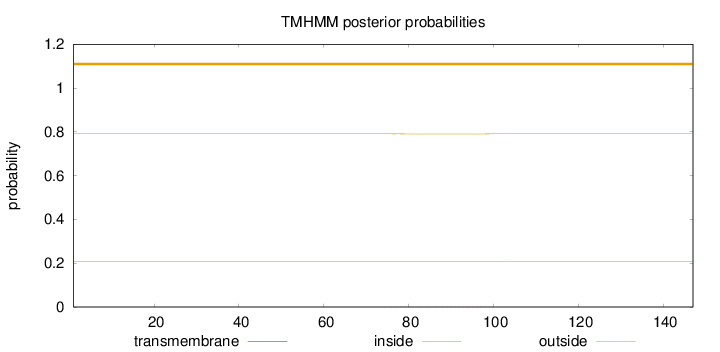
Length:
147
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01229
Exp number, first 60 AAs:
0.00099
Total prob of N-in:
0.20895
outside
1 - 147
Population Genetic Test Statistics
Pi
292.658951
Theta
139.868759
Tajima's D
0.696591
CLR
0.436523
CSRT
0.572571371431428
Interpretation
Uncertain
