Gene
KWMTBOMO03597
Pre Gene Modal
BGIBMGA006317
Annotation
PREDICTED:_CCR4-NOT_transcription_complex_subunit_6_isoform_X1_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.303 PlasmaMembrane Reliability : 1.556
Sequence
CDS
ATGTGGTTAACGAGAGCTGCGTCAGTGTTAAGAGACACGGCCCGATACATAATGAATACTTTATCTGTTGCAGGCATCTTCACGGTGATGTGCTATAACGTGTTGTGCGACAAGTACGCGACAAGACAGATGTACGGCTACTGTCCGAGTTGGGCGCTCGAGTGGGACTACAGGAAGAAGGGTATTTTAGACGAAATTAGGCATTACTCAGCAGACCTTATTAGTTTACAGGAAGTCGAAACGGATCAGTTCTATAATTTTTTCCTGCCAGAATTAAAACAGGACGGTTATGACGGAATTTTCTCGCCGAAATCTCGTGCAAAGACGATGTCAGAATCGGAGAGGAAATACGTCGACGGCTGTGCAATATTTTTCAGATCTGCAAAGTTCGCATTAGTTAAGGAGCACCTCATAGAGTTCAATCAGCTGGCGATGGCCAACAGCGAGGGATCAGACAACATGTTAAATCGAGTAATGCCGAAGGACAACATAGGTCTTGCAGCCTTATTGAAGACTAAAGAAGCCGCCTGGGAGAATGGTAATGTTGCTAGTCTACCAACGGACTCGTCAATGATCGGCCAGCCGATACTCGTCTGCACAGCTCACATCCACTGGGACCCAGAGTTTTGCGACGTGAAACTCATCCAAACGATGATGTTAAGCAACGAACTCAGAACGATCCTGGACGATTCGGGAAGGACATTACGTCTCGCCGGACAGAGAGACAACGTGCAGCTGTTGCTCTGCGGGGACTTCAATTCCTTGCCTGACAGCGGCGTGGTCGAGTTCCTGTCCGCGGGGCGCGTCTCGTCTGAGCACCGCGACTTCAAGGAGCTCGGTTACGCGACGTCCCTGCGCCGCATGCCCGGCTCCGAGCACGAGTTCACGCACAACTTCAAGCTGGCCTCGGCCTACAGCGAGGACATCATGCCCTACACCAACTACACGTTTGACTTCAAAGGTATTATCGACTACATATTCTACAGTAAACAGACGATGACCCCCTTAGGGTTGTTGGGTCCCTTGTCTCAGGAATGGTTCCGCGAGCACAAGGTCGTTGGCTGTCCTCATCCGCACATACCTTCAGATCACTTCCCGCTGCTAGTTGAGCTGGAGATGTCCCCAAGCACGAGCGGCAACTCGAACGGTCTGATCGGCCGCAGGTAG
Protein
MWLTRAASVLRDTARYIMNTLSVAGIFTVMCYNVLCDKYATRQMYGYCPSWALEWDYRKKGILDEIRHYSADLISLQEVETDQFYNFFLPELKQDGYDGIFSPKSRAKTMSESERKYVDGCAIFFRSAKFALVKEHLIEFNQLAMANSEGSDNMLNRVMPKDNIGLAALLKTKEAAWENGNVASLPTDSSMIGQPILVCTAHIHWDPEFCDVKLIQTMMLSNELRTILDDSGRTLRLAGQRDNVQLLLCGDFNSLPDSGVVEFLSAGRVSSEHRDFKELGYATSLRRMPGSEHEFTHNFKLASAYSEDIMPYTNYTFDFKGIIDYIFYSKQTMTPLGLLGPLSQEWFREHKVVGCPHPHIPSDHFPLLVELEMSPSTSGNSNGLIGRR
Summary
Uniprot
H9J9X2
A0A2A4JTR2
A0A2H1WK44
A0A2W1C1F0
A0A194QA83
A0A212ENC7
+ More
A0A0L7LUG5 A0A158NGC5 A0A0C9QBJ1 K7IY09 A0A195F1D6 A0A232FAG4 A0A182M5I9 A0A182QRD3 A0A182PLG6 A0A182RI29 A0A1B6GUG2 A0A3F2YSL0 Q7PHD6 A0A1B6HNF1 A0A1Q3G0G2 A0A1Q3G0C1 A0A1B6M6E9 A0A1B6GN47 A0A182GYR1 A0A2M4CTC6 A0A2M3Z722 A0A2M4AGT2 A0A2M4BRG4 A0A1W7R5K2 A0A2M4ALW3 A0A0P6IV40 A0A182KZT2 A0A182JAP9 A0A182SNL3 A0A1L8DGB7 W5JSW9 Q16KP3 A0A182FAF0 A0A1B6DP66 A0A084W3D9 U5EWB3 A0A1J1J3R7 A0A1A9Y109 A0A034VBG2 A0A0A1X6R6 A0A1B0A1L8 A0A1A9VAG3 W8BNW5 W8CA70 A0A1Y1MNJ0 I5ANY9 A0A0P9A2F2 A0A139WMR9 A0A0L0BYJ6 I5ANZ1 A0A139WMX1 A0A0P9CCF2 A0A1I8NBS3 A0A0K8WGX7 A0A0K8W417 A0A1I8NBR7 A0A1I8NBR4 A0A1Y1MNI4 A0A224XEE1 A0A0P8YPN7 A0A069DZ85 A0A023F4L8 T1HSH1 A0A0Q9WUX3 A0A0Q9X6F8 A0A2J7RS33 A0A0M5J4L3 A0A0Q9WAS8 A0A0Q9WZ05 A0A1B0G523 A0A336MBF0 A0A1W4WVI9 A0A3B0KAI5 A0A1W4US32 A0A3B0JKD7 Q8MTZ5 A0A1W4V5V3 A0A1I8NRN8 Q7K112 Q8IMX1 Q9VCB6 A0A0R1E6E7 A0A3B0KGB6 A0A0R1E5V2 A0A0K8W4M9 J9JWD6 A0A2H8TH81 A0A0Q9VZU6 Q8IMX0 A0A0R1E745 T1IXS6 E9G5I7 A0A0P4Z4Y4 A0A0P5XCF2
A0A0L7LUG5 A0A158NGC5 A0A0C9QBJ1 K7IY09 A0A195F1D6 A0A232FAG4 A0A182M5I9 A0A182QRD3 A0A182PLG6 A0A182RI29 A0A1B6GUG2 A0A3F2YSL0 Q7PHD6 A0A1B6HNF1 A0A1Q3G0G2 A0A1Q3G0C1 A0A1B6M6E9 A0A1B6GN47 A0A182GYR1 A0A2M4CTC6 A0A2M3Z722 A0A2M4AGT2 A0A2M4BRG4 A0A1W7R5K2 A0A2M4ALW3 A0A0P6IV40 A0A182KZT2 A0A182JAP9 A0A182SNL3 A0A1L8DGB7 W5JSW9 Q16KP3 A0A182FAF0 A0A1B6DP66 A0A084W3D9 U5EWB3 A0A1J1J3R7 A0A1A9Y109 A0A034VBG2 A0A0A1X6R6 A0A1B0A1L8 A0A1A9VAG3 W8BNW5 W8CA70 A0A1Y1MNJ0 I5ANY9 A0A0P9A2F2 A0A139WMR9 A0A0L0BYJ6 I5ANZ1 A0A139WMX1 A0A0P9CCF2 A0A1I8NBS3 A0A0K8WGX7 A0A0K8W417 A0A1I8NBR7 A0A1I8NBR4 A0A1Y1MNI4 A0A224XEE1 A0A0P8YPN7 A0A069DZ85 A0A023F4L8 T1HSH1 A0A0Q9WUX3 A0A0Q9X6F8 A0A2J7RS33 A0A0M5J4L3 A0A0Q9WAS8 A0A0Q9WZ05 A0A1B0G523 A0A336MBF0 A0A1W4WVI9 A0A3B0KAI5 A0A1W4US32 A0A3B0JKD7 Q8MTZ5 A0A1W4V5V3 A0A1I8NRN8 Q7K112 Q8IMX1 Q9VCB6 A0A0R1E6E7 A0A3B0KGB6 A0A0R1E5V2 A0A0K8W4M9 J9JWD6 A0A2H8TH81 A0A0Q9VZU6 Q8IMX0 A0A0R1E745 T1IXS6 E9G5I7 A0A0P4Z4Y4 A0A0P5XCF2
Pubmed
19121390
28756777
26354079
22118469
26227816
21347285
+ More
20075255 28648823 12364791 14747013 17210077 26483478 26999592 20966253 20920257 23761445 17510324 24438588 25348373 25830018 24495485 28004739 15632085 17994087 18057021 18362917 19820115 26108605 25315136 26334808 25474469 11747467 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 21292972
20075255 28648823 12364791 14747013 17210077 26483478 26999592 20966253 20920257 23761445 17510324 24438588 25348373 25830018 24495485 28004739 15632085 17994087 18057021 18362917 19820115 26108605 25315136 26334808 25474469 11747467 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 21292972
EMBL
BABH01026016
NWSH01000685
PCG74782.1
ODYU01009196
SOQ53450.1
KZ149893
+ More
PZC78950.1 KQ459249 KPJ02387.1 AGBW02013676 OWR42994.1 JTDY01000061 KOB79100.1 ADTU01014789 ADTU01014790 ADTU01014791 ADTU01014792 ADTU01014793 GBYB01000624 JAG70391.1 KQ981864 KYN34278.1 NNAY01000626 OXU27317.1 AXCM01000106 AXCN02001206 GECZ01003697 JAS66072.1 APCN01005808 AAAB01008898 EAA44583.2 GECU01031503 GECU01012334 JAS76203.1 JAS95372.1 GFDL01001789 JAV33256.1 GFDL01001790 JAV33255.1 GEBQ01008513 JAT31464.1 GECZ01005938 JAS63831.1 JXUM01098067 KQ564392 KXJ72348.1 GGFL01004337 MBW68515.1 GGFM01003539 MBW24290.1 GGFK01006626 MBW39947.1 GGFJ01006453 MBW55594.1 GEHC01001199 JAV46446.1 GGFK01008449 MBW41770.1 GDUN01000328 JAN95591.1 GFDF01008669 JAV05415.1 ADMH02000180 ETN67457.1 CH477947 EAT34872.1 GEDC01009841 JAS27457.1 ATLV01019934 KE525286 KFB44733.1 GANO01000642 JAB59229.1 CVRI01000069 CRL07040.1 GAKP01020064 JAC38888.1 GBXI01014878 GBXI01007954 JAC99413.1 JAD06338.1 GAMC01006028 GAMC01006027 JAC00528.1 GAMC01006026 JAC00530.1 GEZM01026115 JAV87233.1 CM000070 EIM52674.1 CH902617 KPU80718.1 KPU80721.1 KQ971312 KYB29329.1 JRES01001160 KNC25090.1 EIM52676.2 KYB29330.1 KPU80720.1 GDHF01001977 JAI50337.1 GDHF01029958 GDHF01028526 GDHF01015296 GDHF01006689 JAI22356.1 JAI23788.1 JAI37018.1 JAI45625.1 GEZM01026114 JAV87234.1 GFTR01007062 JAW09364.1 KPU80719.1 GBGD01001080 JAC87809.1 GBBI01002440 JAC16272.1 ACPB03008338 CH964232 KRF99336.1 CH933806 KRG00948.1 NEVH01000595 PNF43639.1 CP012526 ALC45836.1 CH940656 KRF78354.1 KRF78356.1 KRG00949.1 CCAG010008343 UFQT01000793 SSX27270.1 OUUW01000007 SPP82686.1 SPP82685.1 AY043267 AAK85705.1 AE014297 AY069458 AAF56258.2 AAF56259.2 AAL39603.1 AAO41599.1 BT010039 AAN13986.1 AAQ22508.1 BT001344 AAF56256.2 AAN71099.1 CM000160 KRK04589.1 KRK04591.1 SPP82688.1 KRK04592.1 GDHF01006258 JAI46056.1 ABLF02032684 ABLF02032685 GFXV01001660 MBW13465.1 KRF78355.1 AAN13987.1 KRK04590.1 JH431661 GL732532 EFX85626.1 GDIP01217910 JAJ05492.1 GDIP01074224 JAM29491.1
PZC78950.1 KQ459249 KPJ02387.1 AGBW02013676 OWR42994.1 JTDY01000061 KOB79100.1 ADTU01014789 ADTU01014790 ADTU01014791 ADTU01014792 ADTU01014793 GBYB01000624 JAG70391.1 KQ981864 KYN34278.1 NNAY01000626 OXU27317.1 AXCM01000106 AXCN02001206 GECZ01003697 JAS66072.1 APCN01005808 AAAB01008898 EAA44583.2 GECU01031503 GECU01012334 JAS76203.1 JAS95372.1 GFDL01001789 JAV33256.1 GFDL01001790 JAV33255.1 GEBQ01008513 JAT31464.1 GECZ01005938 JAS63831.1 JXUM01098067 KQ564392 KXJ72348.1 GGFL01004337 MBW68515.1 GGFM01003539 MBW24290.1 GGFK01006626 MBW39947.1 GGFJ01006453 MBW55594.1 GEHC01001199 JAV46446.1 GGFK01008449 MBW41770.1 GDUN01000328 JAN95591.1 GFDF01008669 JAV05415.1 ADMH02000180 ETN67457.1 CH477947 EAT34872.1 GEDC01009841 JAS27457.1 ATLV01019934 KE525286 KFB44733.1 GANO01000642 JAB59229.1 CVRI01000069 CRL07040.1 GAKP01020064 JAC38888.1 GBXI01014878 GBXI01007954 JAC99413.1 JAD06338.1 GAMC01006028 GAMC01006027 JAC00528.1 GAMC01006026 JAC00530.1 GEZM01026115 JAV87233.1 CM000070 EIM52674.1 CH902617 KPU80718.1 KPU80721.1 KQ971312 KYB29329.1 JRES01001160 KNC25090.1 EIM52676.2 KYB29330.1 KPU80720.1 GDHF01001977 JAI50337.1 GDHF01029958 GDHF01028526 GDHF01015296 GDHF01006689 JAI22356.1 JAI23788.1 JAI37018.1 JAI45625.1 GEZM01026114 JAV87234.1 GFTR01007062 JAW09364.1 KPU80719.1 GBGD01001080 JAC87809.1 GBBI01002440 JAC16272.1 ACPB03008338 CH964232 KRF99336.1 CH933806 KRG00948.1 NEVH01000595 PNF43639.1 CP012526 ALC45836.1 CH940656 KRF78354.1 KRF78356.1 KRG00949.1 CCAG010008343 UFQT01000793 SSX27270.1 OUUW01000007 SPP82686.1 SPP82685.1 AY043267 AAK85705.1 AE014297 AY069458 AAF56258.2 AAF56259.2 AAL39603.1 AAO41599.1 BT010039 AAN13986.1 AAQ22508.1 BT001344 AAF56256.2 AAN71099.1 CM000160 KRK04589.1 KRK04591.1 SPP82688.1 KRK04592.1 GDHF01006258 JAI46056.1 ABLF02032684 ABLF02032685 GFXV01001660 MBW13465.1 KRF78355.1 AAN13987.1 KRK04590.1 JH431661 GL732532 EFX85626.1 GDIP01217910 JAJ05492.1 GDIP01074224 JAM29491.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000037510
UP000005205
+ More
UP000002358 UP000078541 UP000215335 UP000075883 UP000075886 UP000075885 UP000075900 UP000075840 UP000007062 UP000069940 UP000249989 UP000075882 UP000075880 UP000075901 UP000000673 UP000008820 UP000069272 UP000030765 UP000183832 UP000092443 UP000092445 UP000078200 UP000001819 UP000007801 UP000007266 UP000037069 UP000095301 UP000015103 UP000007798 UP000009192 UP000235965 UP000092553 UP000008792 UP000092444 UP000192223 UP000268350 UP000192221 UP000095300 UP000000803 UP000002282 UP000007819 UP000000305
UP000002358 UP000078541 UP000215335 UP000075883 UP000075886 UP000075885 UP000075900 UP000075840 UP000007062 UP000069940 UP000249989 UP000075882 UP000075880 UP000075901 UP000000673 UP000008820 UP000069272 UP000030765 UP000183832 UP000092443 UP000092445 UP000078200 UP000001819 UP000007801 UP000007266 UP000037069 UP000095301 UP000015103 UP000007798 UP000009192 UP000235965 UP000092553 UP000008792 UP000092444 UP000192223 UP000268350 UP000192221 UP000095300 UP000000803 UP000002282 UP000007819 UP000000305
Interpro
SUPFAM
SSF56219
SSF56219
Gene 3D
ProteinModelPortal
H9J9X2
A0A2A4JTR2
A0A2H1WK44
A0A2W1C1F0
A0A194QA83
A0A212ENC7
+ More
A0A0L7LUG5 A0A158NGC5 A0A0C9QBJ1 K7IY09 A0A195F1D6 A0A232FAG4 A0A182M5I9 A0A182QRD3 A0A182PLG6 A0A182RI29 A0A1B6GUG2 A0A3F2YSL0 Q7PHD6 A0A1B6HNF1 A0A1Q3G0G2 A0A1Q3G0C1 A0A1B6M6E9 A0A1B6GN47 A0A182GYR1 A0A2M4CTC6 A0A2M3Z722 A0A2M4AGT2 A0A2M4BRG4 A0A1W7R5K2 A0A2M4ALW3 A0A0P6IV40 A0A182KZT2 A0A182JAP9 A0A182SNL3 A0A1L8DGB7 W5JSW9 Q16KP3 A0A182FAF0 A0A1B6DP66 A0A084W3D9 U5EWB3 A0A1J1J3R7 A0A1A9Y109 A0A034VBG2 A0A0A1X6R6 A0A1B0A1L8 A0A1A9VAG3 W8BNW5 W8CA70 A0A1Y1MNJ0 I5ANY9 A0A0P9A2F2 A0A139WMR9 A0A0L0BYJ6 I5ANZ1 A0A139WMX1 A0A0P9CCF2 A0A1I8NBS3 A0A0K8WGX7 A0A0K8W417 A0A1I8NBR7 A0A1I8NBR4 A0A1Y1MNI4 A0A224XEE1 A0A0P8YPN7 A0A069DZ85 A0A023F4L8 T1HSH1 A0A0Q9WUX3 A0A0Q9X6F8 A0A2J7RS33 A0A0M5J4L3 A0A0Q9WAS8 A0A0Q9WZ05 A0A1B0G523 A0A336MBF0 A0A1W4WVI9 A0A3B0KAI5 A0A1W4US32 A0A3B0JKD7 Q8MTZ5 A0A1W4V5V3 A0A1I8NRN8 Q7K112 Q8IMX1 Q9VCB6 A0A0R1E6E7 A0A3B0KGB6 A0A0R1E5V2 A0A0K8W4M9 J9JWD6 A0A2H8TH81 A0A0Q9VZU6 Q8IMX0 A0A0R1E745 T1IXS6 E9G5I7 A0A0P4Z4Y4 A0A0P5XCF2
A0A0L7LUG5 A0A158NGC5 A0A0C9QBJ1 K7IY09 A0A195F1D6 A0A232FAG4 A0A182M5I9 A0A182QRD3 A0A182PLG6 A0A182RI29 A0A1B6GUG2 A0A3F2YSL0 Q7PHD6 A0A1B6HNF1 A0A1Q3G0G2 A0A1Q3G0C1 A0A1B6M6E9 A0A1B6GN47 A0A182GYR1 A0A2M4CTC6 A0A2M3Z722 A0A2M4AGT2 A0A2M4BRG4 A0A1W7R5K2 A0A2M4ALW3 A0A0P6IV40 A0A182KZT2 A0A182JAP9 A0A182SNL3 A0A1L8DGB7 W5JSW9 Q16KP3 A0A182FAF0 A0A1B6DP66 A0A084W3D9 U5EWB3 A0A1J1J3R7 A0A1A9Y109 A0A034VBG2 A0A0A1X6R6 A0A1B0A1L8 A0A1A9VAG3 W8BNW5 W8CA70 A0A1Y1MNJ0 I5ANY9 A0A0P9A2F2 A0A139WMR9 A0A0L0BYJ6 I5ANZ1 A0A139WMX1 A0A0P9CCF2 A0A1I8NBS3 A0A0K8WGX7 A0A0K8W417 A0A1I8NBR7 A0A1I8NBR4 A0A1Y1MNI4 A0A224XEE1 A0A0P8YPN7 A0A069DZ85 A0A023F4L8 T1HSH1 A0A0Q9WUX3 A0A0Q9X6F8 A0A2J7RS33 A0A0M5J4L3 A0A0Q9WAS8 A0A0Q9WZ05 A0A1B0G523 A0A336MBF0 A0A1W4WVI9 A0A3B0KAI5 A0A1W4US32 A0A3B0JKD7 Q8MTZ5 A0A1W4V5V3 A0A1I8NRN8 Q7K112 Q8IMX1 Q9VCB6 A0A0R1E6E7 A0A3B0KGB6 A0A0R1E5V2 A0A0K8W4M9 J9JWD6 A0A2H8TH81 A0A0Q9VZU6 Q8IMX0 A0A0R1E745 T1IXS6 E9G5I7 A0A0P4Z4Y4 A0A0P5XCF2
PDB
5DV4
E-value=3.10754e-125,
Score=1148
Ontologies
GO
Topology
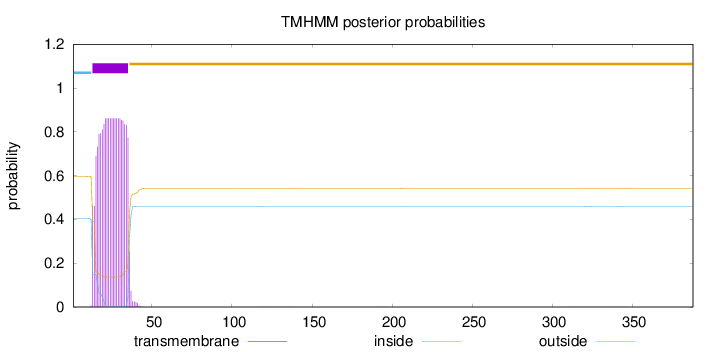
Length:
388
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
18.79313
Exp number, first 60 AAs:
18.76182
Total prob of N-in:
0.40432
POSSIBLE N-term signal
sequence
inside
1 - 12
TMhelix
13 - 35
outside
36 - 388
Population Genetic Test Statistics
Pi
250.781161
Theta
177.474427
Tajima's D
1.223473
CLR
0.458762
CSRT
0.722813859307035
Interpretation
Uncertain
