Gene
KWMTBOMO03588
Pre Gene Modal
BGIBMGA006278
Annotation
PREDICTED:_transmembrane_protein_135-like_isoform_X1_[Papilio_machaon]
Location in the cell
PlasmaMembrane Reliability : 2.835
Sequence
CDS
ATGCTCCGTGCAGTAATACCTAAAATCCTGACTCCGATGAAAGCGTTGAAGTCCCTAAGACTGGCCCATTTGAGACTGGGAATTTTCTTCGGAGGCTACATAGGAATATATAGGCTGGTGGTTTGTGCACTGTGTCGAGCGTATGGCAGGGACAGCGCTTCTTACGCGTTGCCCGCGGGCCTCCTCGCCGGCGCCGCGTTCAGGGCGGCCCCCTCCACGCCGATCGCGTTGGCCCCGATCACGTCCTCCCTGCAAATCTTGGCGTCGTGGGCGTATCAGCGCGAGATGTTACCGGCGAACCTGCCGCTGGTCGAGATACTGTATTGCTTGTGTCAGGGGCTGTTGTTCCACGCTCGCGTCATGCACGAGGACGCCTGCCCCCGGTACATCATCAACCTGATGCACACCGTCACCAGCTCTAAGGCCGATCAAATTCAAGGTGCTTTCATAGAGAAAATGTTGAAGAGCTTGTGA
Protein
MLRAVIPKILTPMKALKSLRLAHLRLGIFFGGYIGIYRLVVCALCRAYGRDSASYALPAGLLAGAAFRAAPSTPIALAPITSSLQILASWAYQREMLPANLPLVEILYCLCQGLLFHARVMHEDACPRYIINLMHTVTSSKADQIQGAFIEKMLKSL
Summary
Uniprot
EMBL
BABH01025997
KZ149893
PZC78954.1
NWSH01000409
PCG76607.1
PCG76605.1
+ More
GDQN01011660 JAT79394.1 ODYU01010110 SOQ54984.1 KQ460647 KPJ13199.1 AGBW02009470 OWR50748.1 KQ459249 KPJ02390.1 PCG76606.1 AGBW02009711 OWR50051.1 KQ460170 KPJ17226.1 ODYU01006255 SOQ47928.1 NWSH01000391 PCG76737.1 PZC78908.1 GDQN01010648 GDQN01008315 GDQN01007732 JAT80406.1 JAT82739.1 JAT83322.1 KQ459011 KPJ04357.1 AGBW02009622 OWR50303.1 BABH01026127
GDQN01011660 JAT79394.1 ODYU01010110 SOQ54984.1 KQ460647 KPJ13199.1 AGBW02009470 OWR50748.1 KQ459249 KPJ02390.1 PCG76606.1 AGBW02009711 OWR50051.1 KQ460170 KPJ17226.1 ODYU01006255 SOQ47928.1 NWSH01000391 PCG76737.1 PZC78908.1 GDQN01010648 GDQN01008315 GDQN01007732 JAT80406.1 JAT82739.1 JAT83322.1 KQ459011 KPJ04357.1 AGBW02009622 OWR50303.1 BABH01026127
Proteomes
Pfam
PF15982 TMEM135_C_rich
ProteinModelPortal
Ontologies
GO
PANTHER
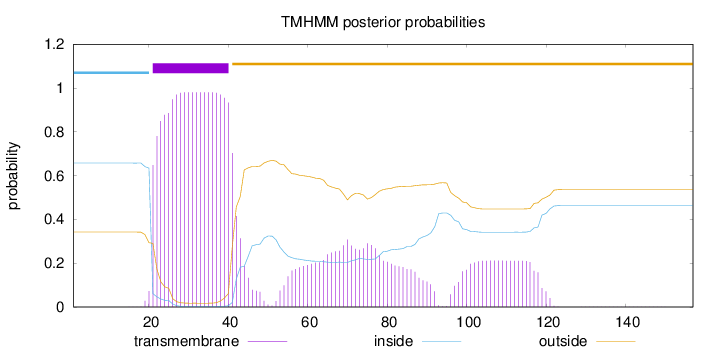
Topology
Length:
157
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
33.2486
Exp number, first 60 AAs:
21.97374
Total prob of N-in:
0.65743
POSSIBLE N-term signal
sequence
inside
1 - 20
TMhelix
21 - 40
outside
41 - 157
Population Genetic Test Statistics
Pi
202.628702
Theta
130.354627
Tajima's D
0.985128
CLR
0.337384
CSRT
0.658667066646668
Interpretation
Uncertain
