Gene
KWMTBOMO03582
Pre Gene Modal
BGIBMGA011165
Annotation
PREDICTED:_uncharacterized_protein_LOC106709547_[Papilio_machaon]
Full name
Galactose-1-phosphate uridylyltransferase
Location in the cell
Mitochondrial Reliability : 1.547 Nuclear Reliability : 1.797
Sequence
CDS
ATGTCTCAAGAAATCTACAGAGTTGGTGTGCGCATTCCGCCATTCTGCTCAGAGGACCCAGAAATGTGGTTTAGTCAGATAGAGATGCAATTTAAGCTCTCAGGGATATCGGTGGACGATACTAAGTTTACGTATGTGTCAGCGAACCTGGAACCACAATATGCCAAGGAGGTGAGAGACATCATAACTAAACCGCCAGCTTCAAATAAATACGAAAAACTCAAAACAGAGCTGATCAAGCGACTATCTATATCACAGGAGAAGCGTGTAAGACAACTTCTCATGCACGAAGAACTCGGAGATCGTAAACCGTCTCAGTTTCTACGCCACTTGACAAGTCTAGCTGGTTCAGACGTTCCATCTGATTTTATAATAACCATCTGGACCAGTCGTCTGCCTGTGAACGTGCAAACAGCAGTCGCTACGATTAAAGGATCGCCAATGGAAAAGCTGAGTGACTTGGCAGATGTCGTATACGAAATGGCCCCAGCAGTACCACAGGTGGCAAGCGCGTCTTCCCAAGCCCAAGGCAATGAGCAAGTCAATCCGACACTGACCATGCAGTTTCGCGTCGGAAAACTCCTGGGGCGGTCGGAAGTAGCGGCCAACGACTGCCCACATTCTTCAGGCCGTCTTTTTATCACCGACCGTACTTCAAAAATGCAGTTCCTAGTGGATACTGGTTCCGATCTGTGTGTTTTTCCACGGTCGGCTGTGAAGGCGCCCTGTACCAAGTCGAAATATGATCTCGTAGCCGCGAATAATTCTATTATACACACTTATGGCCCTGTAACTCTTCATTTGGACTTTGGACTAAGAAGAGACTTCGCGTGGAAATTCACTGTTGCAGACGTATCTAAGCCCATAATCGGAGTGGATTTCTTGTCTTACTACAACTTACTCGTCGATTGCAGAAAACAGCGTATCATTGATGGCATCACTTCTCTGTCTGCAGTCGCGACACCTATCGGGATGACTAAAGACCTTCCTTCTGTCAAAGCCGTCACTGGTGAAACTGACTTCCACGATATTCTGCGAGAGTTCCCAGAAATCACAAGGCCAGCAGGTATGCTCTCTACCACATTTAAGCATAACACCGTTCATCATATTAGAACTACACCGGGACCTCCCGTTGCTTGCAAGCCTCGACGTTTGGATCCAGCACGTATGCAAGCCGCGAAGAAAGAGTTCGAGGATATGTTGGCAAGCGGCGTAGCCCGGCGTTCAGAGAGTGCATGGTCTTCTGCTCTGCACTTGGTGAAGAAGAAATATGATGGATGGCGTCCTTGTGGCGATTACCGATCACTGAATGCGAGGACGATACCTGATAGGTATCCAATACGCCACATCCATGACTTCTCACAACAACTTTCTGGATGTTCCATCTTTTCTAAAATTGATTTGGTGAAGGCTTATCATCAGATAGCCGTTAATCCTGAGGATATACCTAAAACAGCCATCACTACTCCTTTTGGACTTTACGAATTTCCATTTATGACCTTTGGTTTGAGGAATGCCGCTCAGACATTCCAAAGGTTCATAGATGAGGTGCTACTAGGCTTCGACTTCACCTATGGATATATTGATGATATATTATTGTTTTCTTCATCGCTAGAAGAACATAAGCGTCATCTACGTCAACTCTTTGATCGTCTCCGGCAGTATGGCATTTTGGTGAACACAACAAAATGTACTTTTGGTCAATCGGTCGTGGAATTCCTAGGATATGAAGTATCTGCTACAGGTACTAAACCATTGGACACTAAAGTCCAAGCTATCCAGGAATTTGCTGCTCCCAAGACTGTGAAAGAACTCCGAAGATTCTTAGGCATGCTTAATTTCTACCGACGTTTCATACCTAATGCCGCCCAGTTACAAGCTCCGTTGAACAACGCTCTGGCAGGAGTTAAAATGAAAGGTTCACATCCTGTGGATATGGATACAGTCATGTTAAAAGCCTTTGAGGATTGTAAGACTTCTTTATCTCAGGCAACGCTTTTGTCTCACCCGGATCCAACCGCCGATCTTGCGATTGTCACTGACGCGTCTCTCACAGCGATCGGAGCTGTTCTACAACAAAGGAGAGGTGAACATTGGGAACCACTCGCTTTCTACTCCCACAAGCTTAGTTCCACCCAATCGAAGTACAGTCCATATGATCGAGAATTACTGTCAGTTTATGAGGCAATAAAGCATTTTCGACACATGATCGAAGCAAGAAATTTCACCATCTTCACTGATCATAAACCACTGACTTTTGCCTTCTCAGTAAACAGGGAGAAATGTTCACCAAGGCAATTCAGATATCTCGACTACATATCGCAGTTTACTACGGATATTAAATATATACCTGGAAAGGATAACGTAGTCGCCGACGCACTTTCTCGGGTGGATGAAGTCGCTCCATGTATCGACTATGAACGTCTTGCACGGTCTCAGGAACAAGATCCAGAGCTGCAAGATCTTCTGCGTAATGGAAGTTCTCTAAAATTAGAAAGTTATCTTCTGAGTATAGTAGCTTACCCGTTTATTGCGACACCAGTGTGCACCCTCCACGACCATTCATCACAAAAGATATGCGAAGACAAGTCTTCAACACACTGCATGGACTTCATCACCCAGGATCGTCAGCTACAGTCCGACTGGTACTGCCTCACTGTTATTGACCGCTTCACCCGATGGCCTGAGGCGATACCACTCAGCGAAATTACAGCTGAAGCTTGTGCTGCCGCCCTCGTTACCGGCTGGATTGCCAGATTTGGATGTCCACTGAGGGTGACTACCGACCGTGGTCGTCAATTCGAGAGCCACTTATTCAAAGCCATAGCTGCATTAATAGGTGCTCACCACTTCCGGACTACCGCATATCACCCCGCCGCCAACGGAATAGTCGAACGAATGCATCGCCAGTTGAAGGCAGCCATCATGTGCCATACATCTTCGCAATGGACTGAAGCACTTCCTCTAGTACTGATGGGCATGCGCAGTTCCTGGAAGGATGACATTGACGCATCTCCTGCAGAGCTGGTGTACGGCGAGCCCCTCTGTCTGCCAGGTCAATTCATTTCACCAAATGAAGATTACAAAGTGTCTGACGTCACCCAGTATGCAACTCGTCTACGTGCATGCATGGCGAATTTGACACCGAGACCGACATCCTGGCATAGCAATTCACCATTTTATATACCTCGCGATTTACATACAACCTCCCATGTCTTCATCCGACAAGATCATGTGCGTGGTTCTCTGGTACCACCATATGCTGGTCCCTTCAAGGTTGTCAAAAGGCAGCCAAAATTTTTCACTGTCGAAGTTCGAGGGAAGCTTGTAAATGTCTCCATAGATCGTCTGAAGCCGGCCTACATGACGAGAGTCCCAGAAGGTGTGTCTCCAACCAAGCCTGTTGAAAAGACTACAAGAAGCGGCCGAAAGATTAAACTGCCTGATTACTATCGACCGTAG
Protein
MSQEIYRVGVRIPPFCSEDPEMWFSQIEMQFKLSGISVDDTKFTYVSANLEPQYAKEVRDIITKPPASNKYEKLKTELIKRLSISQEKRVRQLLMHEELGDRKPSQFLRHLTSLAGSDVPSDFIITIWTSRLPVNVQTAVATIKGSPMEKLSDLADVVYEMAPAVPQVASASSQAQGNEQVNPTLTMQFRVGKLLGRSEVAANDCPHSSGRLFITDRTSKMQFLVDTGSDLCVFPRSAVKAPCTKSKYDLVAANNSIIHTYGPVTLHLDFGLRRDFAWKFTVADVSKPIIGVDFLSYYNLLVDCRKQRIIDGITSLSAVATPIGMTKDLPSVKAVTGETDFHDILREFPEITRPAGMLSTTFKHNTVHHIRTTPGPPVACKPRRLDPARMQAAKKEFEDMLASGVARRSESAWSSALHLVKKKYDGWRPCGDYRSLNARTIPDRYPIRHIHDFSQQLSGCSIFSKIDLVKAYHQIAVNPEDIPKTAITTPFGLYEFPFMTFGLRNAAQTFQRFIDEVLLGFDFTYGYIDDILLFSSSLEEHKRHLRQLFDRLRQYGILVNTTKCTFGQSVVEFLGYEVSATGTKPLDTKVQAIQEFAAPKTVKELRRFLGMLNFYRRFIPNAAQLQAPLNNALAGVKMKGSHPVDMDTVMLKAFEDCKTSLSQATLLSHPDPTADLAIVTDASLTAIGAVLQQRRGEHWEPLAFYSHKLSSTQSKYSPYDRELLSVYEAIKHFRHMIEARNFTIFTDHKPLTFAFSVNREKCSPRQFRYLDYISQFTTDIKYIPGKDNVVADALSRVDEVAPCIDYERLARSQEQDPELQDLLRNGSSLKLESYLLSIVAYPFIATPVCTLHDHSSQKICEDKSSTHCMDFITQDRQLQSDWYCLTVIDRFTRWPEAIPLSEITAEACAAALVTGWIARFGCPLRVTTDRGRQFESHLFKAIAALIGAHHFRTTAYHPAANGIVERMHRQLKAAIMCHTSSQWTEALPLVLMGMRSSWKDDIDASPAELVYGEPLCLPGQFISPNEDYKVSDVTQYATRLRACMANLTPRPTSWHSNSPFYIPRDLHTTSHVFIRQDHVRGSLVPPYAGPFKVVKRQPKFFTVEVRGKLVNVSIDRLKPAYMTRVPEGVSPTKPVEKTTRSGRKIKLPDYYRP
Summary
Catalytic Activity
alpha-D-galactose 1-phosphate + UDP-alpha-D-glucose = alpha-D-glucose 1-phosphate + UDP-alpha-D-galactose
Similarity
Belongs to the galactose-1-phosphate uridylyltransferase type 1 family.
Uniprot
W4XB88
A0A0A1WEZ6
W4XB89
A0A0J7N8Y6
A0A147BM81
A0A0J7K8C3
+ More
A0A0V1F272 A0A1Y1N1G2 A0A0V1FLR3 J9K9L2 J9KB96 A0A085MC24 A0A2G8JGE7 A0A0J7NWH7 A0A085N4U3 A0A1X7VKM7 K7J7W8 W4Y5A8 A0A2G8L6X1 A0A0J7K7I7 K7J6E7 A0A0V1IJE9 A0A0V1DUC2 A0A0V1NU35 A0A085MZL4 A0A0V1NU14 K7JBA7 A0A085MDY5 D6WTN9 A0A085N387 A0A0C2NDW5 A0A085MR14 A0A085N6R2 A0A085N474 A0A085N6P6 A0A085LN63 A0A0V0XDP3 A0A2G8LS75 A0A0V0TCH2 W4Y2D0 X1XU27 Q4SV63 J9L4R1 A0A0V1KEE1 A0A0V0WBM6 A0A0V0W0Q6 A0A0V0WL88 X1XPJ2 A0A085MUI0 A0A085MDV5 X1XU40
A0A0V1F272 A0A1Y1N1G2 A0A0V1FLR3 J9K9L2 J9KB96 A0A085MC24 A0A2G8JGE7 A0A0J7NWH7 A0A085N4U3 A0A1X7VKM7 K7J7W8 W4Y5A8 A0A2G8L6X1 A0A0J7K7I7 K7J6E7 A0A0V1IJE9 A0A0V1DUC2 A0A0V1NU35 A0A085MZL4 A0A0V1NU14 K7JBA7 A0A085MDY5 D6WTN9 A0A085N387 A0A0C2NDW5 A0A085MR14 A0A085N6R2 A0A085N474 A0A085N6P6 A0A085LN63 A0A0V0XDP3 A0A2G8LS75 A0A0V0TCH2 W4Y2D0 X1XU27 Q4SV63 J9L4R1 A0A0V1KEE1 A0A0V0WBM6 A0A0V0W0Q6 A0A0V0WL88 X1XPJ2 A0A085MUI0 A0A085MDV5 X1XU40
EC Number
2.7.7.12
EMBL
AAGJ04001742
GBXI01017046
JAC97245.1
LBMM01008151
KMQ89105.1
GEGO01003506
+ More
JAR91898.1 LBMM01012030 KMQ86466.1 JYDR01000002 KRY79251.1 GEZM01017686 JAV90495.1 JYDT01000067 KRY86703.1 ABLF02011086 ABLF02042879 ABLF02030693 ABLF02030697 KL363205 KFD54770.1 MRZV01002078 PIK34788.1 LBMM01001181 KMQ96765.1 KL367555 KFD64489.1 AAZX01004093 AAZX01004778 AAGJ04112098 MRZV01000197 PIK55880.1 LBMM01012567 KMQ86146.1 AAZX01005104 JYDV01000289 KRZ22781.1 JYDR01000355 KRY64563.1 JYDM01000100 KRZ87482.1 KL367590 KFD62660.1 KRZ87481.1 AAZX01024401 KL363200 KFD55431.1 KQ971355 EFA07205.2 KL367564 KFD63933.1 JWZT01001363 KII72172.1 KL367778 KFD59660.1 KL367543 KFD65158.1 KL367558 KFD64270.1 KL367544 KFD65142.1 KL363373 KFD46409.1 JYDU01000460 KRX86127.1 MRZV01000001 PIK63042.1 JYDJ01000353 KRX36608.1 AAGJ04130731 ABLF02057446 ABLF02059301 CAAE01013786 CAF95469.1 ABLF02006595 ABLF02011068 ABLF02055413 JYDV01000002 KRZ45603.1 JYDK01000186 KRX73103.1 JYDK01000369 KRX69330.1 JYDK01000100 KRX76473.1 ABLF02058233 KL367644 KFD60876.1 KFD55401.1 ABLF02040931 ABLF02040938
JAR91898.1 LBMM01012030 KMQ86466.1 JYDR01000002 KRY79251.1 GEZM01017686 JAV90495.1 JYDT01000067 KRY86703.1 ABLF02011086 ABLF02042879 ABLF02030693 ABLF02030697 KL363205 KFD54770.1 MRZV01002078 PIK34788.1 LBMM01001181 KMQ96765.1 KL367555 KFD64489.1 AAZX01004093 AAZX01004778 AAGJ04112098 MRZV01000197 PIK55880.1 LBMM01012567 KMQ86146.1 AAZX01005104 JYDV01000289 KRZ22781.1 JYDR01000355 KRY64563.1 JYDM01000100 KRZ87482.1 KL367590 KFD62660.1 KRZ87481.1 AAZX01024401 KL363200 KFD55431.1 KQ971355 EFA07205.2 KL367564 KFD63933.1 JWZT01001363 KII72172.1 KL367778 KFD59660.1 KL367543 KFD65158.1 KL367558 KFD64270.1 KL367544 KFD65142.1 KL363373 KFD46409.1 JYDU01000460 KRX86127.1 MRZV01000001 PIK63042.1 JYDJ01000353 KRX36608.1 AAGJ04130731 ABLF02057446 ABLF02059301 CAAE01013786 CAF95469.1 ABLF02006595 ABLF02011068 ABLF02055413 JYDV01000002 KRZ45603.1 JYDK01000186 KRX73103.1 JYDK01000369 KRX69330.1 JYDK01000100 KRX76473.1 ABLF02058233 KL367644 KFD60876.1 KFD55401.1 ABLF02040931 ABLF02040938
Proteomes
Pfam
Interpro
IPR001584
Integrase_cat-core
+ More
IPR012337 RNaseH-like_sf
IPR000477 RT_dom
IPR041373 RT_RNaseH
IPR036397 RNaseH_sf
IPR041588 Integrase_H2C2
IPR021109 Peptidase_aspartic_dom_sf
IPR001995 Peptidase_A2_cat
IPR041577 RT_RNaseH_2
IPR001969 Aspartic_peptidase_AS
IPR034132 RP_Saci-like
IPR005849 GalP_Utransf_N
IPR019779 GalP_UDPtransf1_His-AS
IPR001937 GalP_UDPtransf1
IPR036691 Endo/exonu/phosph_ase_sf
IPR036265 HIT-like_sf
IPR018061 Retropepsins
IPR012337 RNaseH-like_sf
IPR000477 RT_dom
IPR041373 RT_RNaseH
IPR036397 RNaseH_sf
IPR041588 Integrase_H2C2
IPR021109 Peptidase_aspartic_dom_sf
IPR001995 Peptidase_A2_cat
IPR041577 RT_RNaseH_2
IPR001969 Aspartic_peptidase_AS
IPR034132 RP_Saci-like
IPR005849 GalP_Utransf_N
IPR019779 GalP_UDPtransf1_His-AS
IPR001937 GalP_UDPtransf1
IPR036691 Endo/exonu/phosph_ase_sf
IPR036265 HIT-like_sf
IPR018061 Retropepsins
Gene 3D
ProteinModelPortal
W4XB88
A0A0A1WEZ6
W4XB89
A0A0J7N8Y6
A0A147BM81
A0A0J7K8C3
+ More
A0A0V1F272 A0A1Y1N1G2 A0A0V1FLR3 J9K9L2 J9KB96 A0A085MC24 A0A2G8JGE7 A0A0J7NWH7 A0A085N4U3 A0A1X7VKM7 K7J7W8 W4Y5A8 A0A2G8L6X1 A0A0J7K7I7 K7J6E7 A0A0V1IJE9 A0A0V1DUC2 A0A0V1NU35 A0A085MZL4 A0A0V1NU14 K7JBA7 A0A085MDY5 D6WTN9 A0A085N387 A0A0C2NDW5 A0A085MR14 A0A085N6R2 A0A085N474 A0A085N6P6 A0A085LN63 A0A0V0XDP3 A0A2G8LS75 A0A0V0TCH2 W4Y2D0 X1XU27 Q4SV63 J9L4R1 A0A0V1KEE1 A0A0V0WBM6 A0A0V0W0Q6 A0A0V0WL88 X1XPJ2 A0A085MUI0 A0A085MDV5 X1XU40
A0A0V1F272 A0A1Y1N1G2 A0A0V1FLR3 J9K9L2 J9KB96 A0A085MC24 A0A2G8JGE7 A0A0J7NWH7 A0A085N4U3 A0A1X7VKM7 K7J7W8 W4Y5A8 A0A2G8L6X1 A0A0J7K7I7 K7J6E7 A0A0V1IJE9 A0A0V1DUC2 A0A0V1NU35 A0A085MZL4 A0A0V1NU14 K7JBA7 A0A085MDY5 D6WTN9 A0A085N387 A0A0C2NDW5 A0A085MR14 A0A085N6R2 A0A085N474 A0A085N6P6 A0A085LN63 A0A0V0XDP3 A0A2G8LS75 A0A0V0TCH2 W4Y2D0 X1XU27 Q4SV63 J9L4R1 A0A0V1KEE1 A0A0V0WBM6 A0A0V0W0Q6 A0A0V0WL88 X1XPJ2 A0A085MUI0 A0A085MDV5 X1XU40
PDB
4OL8
E-value=2.51455e-65,
Score=636
Ontologies
GO
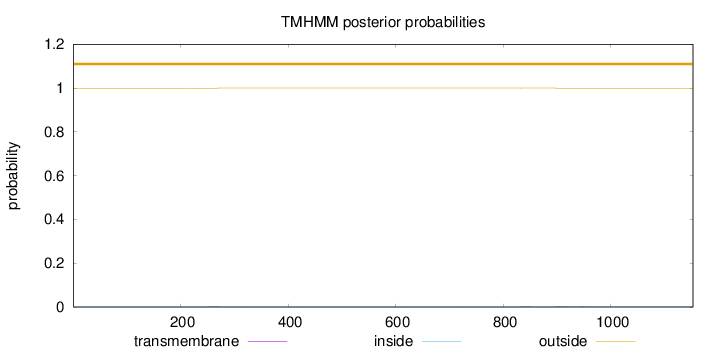
Topology
Length:
1153
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.03928
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00055
outside
1 - 1153
Population Genetic Test Statistics
Pi
158.803767
Theta
154.427968
Tajima's D
0.888117
CLR
0.000172
CSRT
0.629968501574921
Interpretation
Uncertain
