Gene
KWMTBOMO03579
Annotation
putative_Sodium/potassium-transporting_ATPase_subunit_beta-1_[Operophtera_brumata]
Location in the cell
Mitochondrial Reliability : 1.55
Sequence
CDS
ATGGTGAATATTAAAGATAAATATGAAAACCCATCAACCTCGCAGAGTGCTCCAACAACCCCGAGACCTTTGCGTCGAGACCCATCACTTCCAACGTCTTTACCTACATACTCAAAGCCAGATAAACTGCAAGTTGTATCTCAAGAGAAGCCTATATCTAGTAGAAGAAGGTTCCTCAAATACCTTTACAATAGAGAAAAAAAGACATTTTGTGGTAGAACGTGTAAAAATTGGTTTTCGATCATCATCTATTCTGTCCTATATGTAATATCTTTGGCAGCTTATTCCCTGTTATTTTTATATGGTACTTTAATAATATTGAAGGAATTAATTGATTATCGAGTGATGGACAAAGTGGAGTTACTGACTTATTCAGAAAATGGGATCGGCCTTTCAGGGACGCCAGCTTCTGAAGGTGATCAGCCAATAATCTGGTATAGGCAAGGTATTGAAGATGATTACAAAAAATACATTAACGCAATAGACAGACTTTTAATCAAGAATAGAAGGAAAAGGGAGCTCAGTGATTTAGGGCCTTGTGGCGAAGCTCCTTACGGATACGGAGACAAACCTTGTGTGATAATCAAGATAAATAAACAGTTGTATTGGAAAGGGAAACCATTGGACACCAATTCATCTGAAACGAAATTGGCACCAAAGCAAGTTCTGAATTGGATACAGAGAGACAAGAAACTCTGGAATTTCTGCGAAGGGGTTACCTCGTTCGACAAAGAACACTTGAGGGATATCGATTACTATCCAAACCCTCCAGGCATAGATCCTGGCATATTTCCAATAGATTTGCAAAGCGCTTTGCCTGTAGTTGCCATACAAATATCGAATTTTACTATAGGTACATCATTGGCGATAGAGTGTAAACTTTGGTACGAAGGAGGTCCTTCATCTTTTGGGTTTGTAATATATGTTGCTCCAGATAGTCAAGTAGTTTATAGGAAGACGATTGATAGTGAAACAAATAGTGGATGA
Protein
MVNIKDKYENPSTSQSAPTTPRPLRRDPSLPTSLPTYSKPDKLQVVSQEKPISSRRRFLKYLYNREKKTFCGRTCKNWFSIIIYSVLYVISLAAYSLLFLYGTLIILKELIDYRVMDKVELLTYSENGIGLSGTPASEGDQPIIWYRQGIEDDYKKYINAIDRLLIKNRRKRELSDLGPCGEAPYGYGDKPCVIIKINKQLYWKGKPLDTNSSETKLAPKQVLNWIQRDKKLWNFCEGVTSFDKEHLRDIDYYPNPPGIDPGIFPIDLQSALPVVAIQISNFTIGTSLAIECKLWYEGGPSSFGFVIYVAPDSQVVYRKTIDSETNSG
Summary
Uniprot
A0A0L7LI47
A0A2H1WMY9
A0A2A4K715
A0A212EHN7
A0A195CRY7
A0A151WKW9
+ More
A0A151JYH6 A0A195E3L3 A0A151I0X5 A0A158NW52 A0A026WA56 A0A088A4B1 A0A154NZQ9 A0A212F3C2 N6UJ87 A0A1B6FQY1 A0A182QHA3 A0A1J1IHX3 E2B7V1 A0A0M8ZWF1 A0A1J1HFD2 A0A0V0G6C0 A0A194R626 A0A2H1VA10 T1G2E8 A0A182L2K0 A0A1S4GW98 A0A182URH5 A0A182X1I1 A0A2W1BP20 A0A182JE55 A0A0K8TRE2 T1EB20 A0A084VI42 A0A386IQA0 D6WKU1 A0A212F3C0 A0A1B6J844 A0A2M3Z1Y5 A0A182FNA7 J9EAN8 A0A2M4CPQ7 A0A2M4CPK3 Q7Q3B0 A0A2M4BU32 A0A1D2N9I5 A0A2M4AFG7 A0A2C9H707 A0A023EQW5 W5UB64 T1D4D1 B3N5X2 A0A3L8DM08 B4LRJ8 A0A023F2Q2 A0A182HDG2 A0A2A3E7J6 A0A0A9W4G9 A0A1Q3FIL8 B0WIC5 A0A182XZM8 A0A069DSA5 U4U4L8 B4JPW9 A0A1W4UY15 A0A194R6X6 B4JB93 E0VVM0 T1ILP1 T1L398 A0A1I8P7X3 D2A6B8 A0A1B6HRM5 D6WJI8 A0A0P4VU23 R4FNJ2 A0A1B6KSW6 A0A182XZM9 Q16TS1 A0A1W4WPD8 A0A0N5BNP4 A0A2I9LP91 W5JUY6 A0A067RAH3 A0A226DN85 B3MKP8 A0A1Y1LAS0 R7U886 B4P6W4 A0A0V0G820 A0A182PJP0 A0A023ENE1
A0A151JYH6 A0A195E3L3 A0A151I0X5 A0A158NW52 A0A026WA56 A0A088A4B1 A0A154NZQ9 A0A212F3C2 N6UJ87 A0A1B6FQY1 A0A182QHA3 A0A1J1IHX3 E2B7V1 A0A0M8ZWF1 A0A1J1HFD2 A0A0V0G6C0 A0A194R626 A0A2H1VA10 T1G2E8 A0A182L2K0 A0A1S4GW98 A0A182URH5 A0A182X1I1 A0A2W1BP20 A0A182JE55 A0A0K8TRE2 T1EB20 A0A084VI42 A0A386IQA0 D6WKU1 A0A212F3C0 A0A1B6J844 A0A2M3Z1Y5 A0A182FNA7 J9EAN8 A0A2M4CPQ7 A0A2M4CPK3 Q7Q3B0 A0A2M4BU32 A0A1D2N9I5 A0A2M4AFG7 A0A2C9H707 A0A023EQW5 W5UB64 T1D4D1 B3N5X2 A0A3L8DM08 B4LRJ8 A0A023F2Q2 A0A182HDG2 A0A2A3E7J6 A0A0A9W4G9 A0A1Q3FIL8 B0WIC5 A0A182XZM8 A0A069DSA5 U4U4L8 B4JPW9 A0A1W4UY15 A0A194R6X6 B4JB93 E0VVM0 T1ILP1 T1L398 A0A1I8P7X3 D2A6B8 A0A1B6HRM5 D6WJI8 A0A0P4VU23 R4FNJ2 A0A1B6KSW6 A0A182XZM9 Q16TS1 A0A1W4WPD8 A0A0N5BNP4 A0A2I9LP91 W5JUY6 A0A067RAH3 A0A226DN85 B3MKP8 A0A1Y1LAS0 R7U886 B4P6W4 A0A0V0G820 A0A182PJP0 A0A023ENE1
Pubmed
26227816
22118469
21347285
24508170
23537049
20798317
+ More
26354079 23254933 20966253 12364791 28756777 26369729 24438588 30245637 18362917 19820115 17510324 14747013 17210077 27289101 24945155 24330624 17994087 30249741 25474469 26483478 25401762 26823975 25244985 26334808 20566863 27129103 29248469 20920257 23761445 24845553 28004739 17550304
26354079 23254933 20966253 12364791 28756777 26369729 24438588 30245637 18362917 19820115 17510324 14747013 17210077 27289101 24945155 24330624 17994087 30249741 25474469 26483478 25401762 26823975 25244985 26334808 20566863 27129103 29248469 20920257 23761445 24845553 28004739 17550304
EMBL
JTDY01001091
KOB74906.1
ODYU01009413
SOQ53824.1
NWSH01000077
PCG79869.1
+ More
AGBW02014861 OWR40980.1 KQ977329 KYN03503.1 KQ982988 KYQ48529.1 KQ981477 KYN41440.1 KQ979701 KYN19517.1 KQ976606 KYM79323.1 ADTU01027808 ADTU01027809 KK107364 EZA51909.1 KQ434782 KZC04574.1 AGBW02010580 OWR48238.1 APGK01032726 APGK01032727 KB740848 ENN78697.1 GECZ01017189 JAS52580.1 AXCN02002071 AXCN02002072 CVRI01000053 CRK99800.1 GL446209 EFN88244.1 KQ435840 KOX71436.1 CVRI01000001 CRK86547.1 GECL01002590 JAP03534.1 KQ460644 KPJ13273.1 ODYU01001458 SOQ37683.1 AMQM01003521 KB096183 ESO07774.1 AAAB01008964 KZ150105 PZC73423.1 GDAI01000897 JAI16706.1 GAMD01000463 JAB01128.1 ATLV01013275 ATLV01013276 ATLV01013277 ATLV01013278 KE524849 KFB37636.1 MG767305 AYD59983.1 KQ971342 EFA02978.1 OWR48239.1 GECU01012359 JAS95347.1 GGFM01001775 MBW22526.1 CH477641 EJY57851.1 GGFL01003105 MBW67283.1 GGFL01003076 MBW67254.1 EAA12801.3 GGFJ01007372 MBW56513.1 LJIJ01000145 ODN01656.1 GGFK01006218 MBW39539.1 GAPW01002579 JAC11019.1 KF813098 AHH35011.1 GALA01001003 JAA93849.1 CH954177 EDV59131.1 QOIP01000007 RLU20858.1 CH940649 EDW63593.1 GBBI01003398 JAC15314.1 JXUM01034452 JXUM01034453 JXUM01034454 JXUM01034455 JXUM01034456 KQ561023 KXJ79990.1 KZ288344 PBC27678.1 GBHO01040920 GBRD01000570 GDHC01017877 JAG02684.1 JAG65251.1 JAQ00752.1 GFDL01007628 JAV27417.1 DS231946 EDS28370.1 GBGD01002278 JAC86611.1 KB631641 ERL84900.1 CH916372 EDV98949.1 KPJ13274.1 CH916368 EDW02898.1 DS235812 EEB17426.1 JH430883 CAEY01001014 CAEY01001015 KQ971347 EFA05491.2 GECU01030411 JAS77295.1 EFA03901.1 GDKW01001068 JAI55527.1 GAHY01000563 JAA76947.1 GEBQ01025457 JAT14520.1 EAT37922.1 GFWZ01000214 MBW20204.1 ADMH02000383 ETN66760.1 KK852595 KDR20645.1 LNIX01000015 OXA46560.1 CH902620 EDV31579.1 GEZM01064643 JAV68676.1 AMQN01008936 AMQN01008937 KB304240 ELU02204.1 CM000158 EDW89933.1 GECL01001960 JAP04164.1 JXUM01045994 JXUM01045995 JXUM01045996 GAPW01002671 KQ561458 JAC10927.1 KXJ78507.1
AGBW02014861 OWR40980.1 KQ977329 KYN03503.1 KQ982988 KYQ48529.1 KQ981477 KYN41440.1 KQ979701 KYN19517.1 KQ976606 KYM79323.1 ADTU01027808 ADTU01027809 KK107364 EZA51909.1 KQ434782 KZC04574.1 AGBW02010580 OWR48238.1 APGK01032726 APGK01032727 KB740848 ENN78697.1 GECZ01017189 JAS52580.1 AXCN02002071 AXCN02002072 CVRI01000053 CRK99800.1 GL446209 EFN88244.1 KQ435840 KOX71436.1 CVRI01000001 CRK86547.1 GECL01002590 JAP03534.1 KQ460644 KPJ13273.1 ODYU01001458 SOQ37683.1 AMQM01003521 KB096183 ESO07774.1 AAAB01008964 KZ150105 PZC73423.1 GDAI01000897 JAI16706.1 GAMD01000463 JAB01128.1 ATLV01013275 ATLV01013276 ATLV01013277 ATLV01013278 KE524849 KFB37636.1 MG767305 AYD59983.1 KQ971342 EFA02978.1 OWR48239.1 GECU01012359 JAS95347.1 GGFM01001775 MBW22526.1 CH477641 EJY57851.1 GGFL01003105 MBW67283.1 GGFL01003076 MBW67254.1 EAA12801.3 GGFJ01007372 MBW56513.1 LJIJ01000145 ODN01656.1 GGFK01006218 MBW39539.1 GAPW01002579 JAC11019.1 KF813098 AHH35011.1 GALA01001003 JAA93849.1 CH954177 EDV59131.1 QOIP01000007 RLU20858.1 CH940649 EDW63593.1 GBBI01003398 JAC15314.1 JXUM01034452 JXUM01034453 JXUM01034454 JXUM01034455 JXUM01034456 KQ561023 KXJ79990.1 KZ288344 PBC27678.1 GBHO01040920 GBRD01000570 GDHC01017877 JAG02684.1 JAG65251.1 JAQ00752.1 GFDL01007628 JAV27417.1 DS231946 EDS28370.1 GBGD01002278 JAC86611.1 KB631641 ERL84900.1 CH916372 EDV98949.1 KPJ13274.1 CH916368 EDW02898.1 DS235812 EEB17426.1 JH430883 CAEY01001014 CAEY01001015 KQ971347 EFA05491.2 GECU01030411 JAS77295.1 EFA03901.1 GDKW01001068 JAI55527.1 GAHY01000563 JAA76947.1 GEBQ01025457 JAT14520.1 EAT37922.1 GFWZ01000214 MBW20204.1 ADMH02000383 ETN66760.1 KK852595 KDR20645.1 LNIX01000015 OXA46560.1 CH902620 EDV31579.1 GEZM01064643 JAV68676.1 AMQN01008936 AMQN01008937 KB304240 ELU02204.1 CM000158 EDW89933.1 GECL01001960 JAP04164.1 JXUM01045994 JXUM01045995 JXUM01045996 GAPW01002671 KQ561458 JAC10927.1 KXJ78507.1
Proteomes
UP000037510
UP000218220
UP000007151
UP000078542
UP000075809
UP000078541
+ More
UP000078492 UP000078540 UP000005205 UP000053097 UP000005203 UP000076502 UP000019118 UP000075886 UP000183832 UP000008237 UP000053105 UP000053240 UP000015101 UP000075882 UP000075903 UP000076407 UP000075880 UP000030765 UP000007266 UP000069272 UP000008820 UP000007062 UP000094527 UP000008711 UP000279307 UP000008792 UP000069940 UP000249989 UP000242457 UP000002320 UP000076408 UP000030742 UP000001070 UP000192221 UP000009046 UP000015104 UP000095300 UP000192223 UP000046392 UP000000673 UP000027135 UP000198287 UP000007801 UP000014760 UP000002282 UP000075885
UP000078492 UP000078540 UP000005205 UP000053097 UP000005203 UP000076502 UP000019118 UP000075886 UP000183832 UP000008237 UP000053105 UP000053240 UP000015101 UP000075882 UP000075903 UP000076407 UP000075880 UP000030765 UP000007266 UP000069272 UP000008820 UP000007062 UP000094527 UP000008711 UP000279307 UP000008792 UP000069940 UP000249989 UP000242457 UP000002320 UP000076408 UP000030742 UP000001070 UP000192221 UP000009046 UP000015104 UP000095300 UP000192223 UP000046392 UP000000673 UP000027135 UP000198287 UP000007801 UP000014760 UP000002282 UP000075885
Pfam
PF00287 Na_K-ATPase
Gene 3D
ProteinModelPortal
A0A0L7LI47
A0A2H1WMY9
A0A2A4K715
A0A212EHN7
A0A195CRY7
A0A151WKW9
+ More
A0A151JYH6 A0A195E3L3 A0A151I0X5 A0A158NW52 A0A026WA56 A0A088A4B1 A0A154NZQ9 A0A212F3C2 N6UJ87 A0A1B6FQY1 A0A182QHA3 A0A1J1IHX3 E2B7V1 A0A0M8ZWF1 A0A1J1HFD2 A0A0V0G6C0 A0A194R626 A0A2H1VA10 T1G2E8 A0A182L2K0 A0A1S4GW98 A0A182URH5 A0A182X1I1 A0A2W1BP20 A0A182JE55 A0A0K8TRE2 T1EB20 A0A084VI42 A0A386IQA0 D6WKU1 A0A212F3C0 A0A1B6J844 A0A2M3Z1Y5 A0A182FNA7 J9EAN8 A0A2M4CPQ7 A0A2M4CPK3 Q7Q3B0 A0A2M4BU32 A0A1D2N9I5 A0A2M4AFG7 A0A2C9H707 A0A023EQW5 W5UB64 T1D4D1 B3N5X2 A0A3L8DM08 B4LRJ8 A0A023F2Q2 A0A182HDG2 A0A2A3E7J6 A0A0A9W4G9 A0A1Q3FIL8 B0WIC5 A0A182XZM8 A0A069DSA5 U4U4L8 B4JPW9 A0A1W4UY15 A0A194R6X6 B4JB93 E0VVM0 T1ILP1 T1L398 A0A1I8P7X3 D2A6B8 A0A1B6HRM5 D6WJI8 A0A0P4VU23 R4FNJ2 A0A1B6KSW6 A0A182XZM9 Q16TS1 A0A1W4WPD8 A0A0N5BNP4 A0A2I9LP91 W5JUY6 A0A067RAH3 A0A226DN85 B3MKP8 A0A1Y1LAS0 R7U886 B4P6W4 A0A0V0G820 A0A182PJP0 A0A023ENE1
A0A151JYH6 A0A195E3L3 A0A151I0X5 A0A158NW52 A0A026WA56 A0A088A4B1 A0A154NZQ9 A0A212F3C2 N6UJ87 A0A1B6FQY1 A0A182QHA3 A0A1J1IHX3 E2B7V1 A0A0M8ZWF1 A0A1J1HFD2 A0A0V0G6C0 A0A194R626 A0A2H1VA10 T1G2E8 A0A182L2K0 A0A1S4GW98 A0A182URH5 A0A182X1I1 A0A2W1BP20 A0A182JE55 A0A0K8TRE2 T1EB20 A0A084VI42 A0A386IQA0 D6WKU1 A0A212F3C0 A0A1B6J844 A0A2M3Z1Y5 A0A182FNA7 J9EAN8 A0A2M4CPQ7 A0A2M4CPK3 Q7Q3B0 A0A2M4BU32 A0A1D2N9I5 A0A2M4AFG7 A0A2C9H707 A0A023EQW5 W5UB64 T1D4D1 B3N5X2 A0A3L8DM08 B4LRJ8 A0A023F2Q2 A0A182HDG2 A0A2A3E7J6 A0A0A9W4G9 A0A1Q3FIL8 B0WIC5 A0A182XZM8 A0A069DSA5 U4U4L8 B4JPW9 A0A1W4UY15 A0A194R6X6 B4JB93 E0VVM0 T1ILP1 T1L398 A0A1I8P7X3 D2A6B8 A0A1B6HRM5 D6WJI8 A0A0P4VU23 R4FNJ2 A0A1B6KSW6 A0A182XZM9 Q16TS1 A0A1W4WPD8 A0A0N5BNP4 A0A2I9LP91 W5JUY6 A0A067RAH3 A0A226DN85 B3MKP8 A0A1Y1LAS0 R7U886 B4P6W4 A0A0V0G820 A0A182PJP0 A0A023ENE1
PDB
4XE5
E-value=2.3081e-07,
Score=130
Ontologies
GO
PANTHER
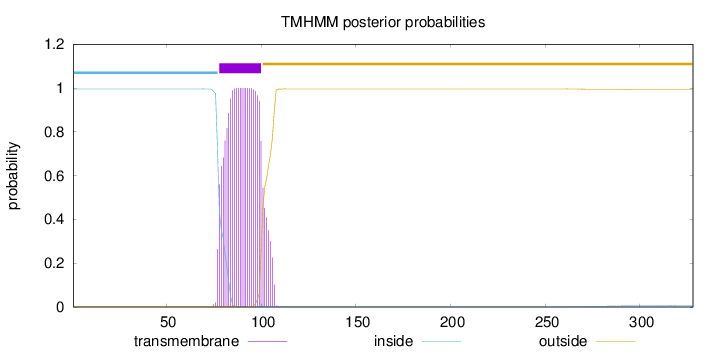
Topology
Length:
328
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
23.68358
Exp number, first 60 AAs:
0
Total prob of N-in:
0.99604
inside
1 - 77
TMhelix
78 - 100
outside
101 - 328
Population Genetic Test Statistics
Pi
314.366261
Theta
224.226301
Tajima's D
1.253887
CLR
0.152763
CSRT
0.731913404329783
Interpretation
Uncertain
