Pre Gene Modal
BGIBMGA006308
Annotation
PREDICTED:_chaoptin_[Bombyx_mori]
Full name
Chaoptin
Alternative Name
Photoreceptor cell-specific membrane protein
Location in the cell
Nuclear Reliability : 2.343
Sequence
CDS
ATGAGAAATAATAATCTGGTGATAGATTTATCTGGTGAAAGAACTAGAAGTTTGATGGCGGTGGTGAAGTTTGGTTACACCCTGGTGGCTGTGACCTTACTGCTGATGATATGGGCGTCGTTGTCCCGAGCTATAGAGCTTCATGTGGACACGGGACATCCGCCATGCTTGTTCCAAGCACTCTGCACTTGTTCAAAACCAGCTGCAGATCTCGGAATCGTGACCTGCAATCACGTGCCCCTTCTGAGGGTCCCCAGTACCATCAACAACTCCAAGGTGTTCATGCTGCAGCTAACTGGAAACAAGATCCGGGATCTAGAGCCACAGTTCTTTCAAACTACAGGACTCTACAGATTAGCGATAAACAAGAACCCTTTGGAAAGTATACACGAAGAGGCCTTTTACGGCTTAGATAATACACTGTGGGAGCTCGAGTTACGTGAAGACAAACTGATTGCAGTACCGAGCAGAGCGTTAAGATATCTGCAGAAGTTACGGTTGTTGGACTTATCAGGCAACGAAATCACGGACATATCAGGCGACAATTGGCGTGGACTAGAGAACACTCTGCAGATATTGATTTTAGCCGACAACTCGATAGCGAACCTACCTTTAGATGCATTTAGTGGACTGCCCTTATTGGAGACACTGGACTTACACGGAAACCACTTGTCTGTTATTGACAGCGGAGTGTTCCGTGATGGAATGAGCAGATTAAATAGGCTGCTATTAGGAGACAACCAGCTGTCGTACATCCCTTATGAGGAACTGTCACCATTAAGGCAACTGAGGGTCTTGGATTTGTCCAGTAACGTTATAAAACAAGTACCACCGTTACATGATCTAAACGGAATCAAACTTTCTTTGGATCTCTTGAAACTAGACAACAATATGATCCAGGTTCTGTATCCGGGCGCATTCAAATACTTTAACGTATTGAACTCGACGACCCTCGACCGGAATCCAATCGTGACGATAAGAGAAGACGCATTCCGTAACGCGAAAATAAAGTCTCTATCGCTAAGAGACTGCAGTATATCCGACCTGTCTCCAGCAGCATTTGCTGGTTTAGAGAACAGTCTGATTAGCTTGGACTTATCCGAGAACAACTTGACAATGGTGTCCAAATTCATGCTGAATAAACTCGATTCTTTAAGGTTCTTAAACTTGAGAGAAAATAAGGTTGACATAAGCTTACTGACCACAAATATGCCGTCGGAGTATCAACAAAGTGCTTTCAACAGTTTCCAATACAGACTTTTTTCGTTAGACGTGAGCGGCGCTAACTCTATAGAAATCAGTCTACAGGATGCTAAGAGAATGAGGTCGTTGAGGTCTCTGTCGGTTGGCAAACTGTTACGTCAAAGCATTAAGACTGAAGATTTCCTGGATTTCGGTATGGAGCTAGAAGACTTGAAGTTATTTGGCAGCAATCTCATCAGGATCGAATCGAGCGCCTTCCAATATGTCAGGACCATCAAGACCCTGGATCTCTCGGAGAATAACATAGAGTACATCGACCCGTTCGCTTTTGCCGAGCTGCACAGCCTTATCACATTGAAGATGGCGCGCGGTTTGTCCGATTCGGTCATAATTCTACCGTTCGAGCCGCTGAAGGCGCTGACCGAACTCCAGTACCTGGACATGAGCAACAACAAACTGAAAAACGTGCCGGACACCTCGTTTCATTTCTTGAAGAACTTGAGATCGTTGAACTTAGAAGATAACGCAATAGAACACTTTTCAAAGGGAACTCTACAAGCCGATATCCACGGAAAATTGGAGAGCATATGCCTCTCCTTGAACCAGATGAAACAAATAGGGCAACACACATTCGTCGAGCTGAAGGAAATGCAAGAAATCCTCATAGAAGATAACCTCATACAGTACATACAGAGGAGAGCGTTCGCCAGCCTCGATAACTTGAAAATTATCAAGCTCAAAGGGAATCGAATCAACGAAATTTCGGAGGAAGCATTCCAAAACATACCGGCTCTCAAAGAGCTTGACGTCTCGTTCAACAAACTGGAAACGTTTAAGTTCTCTATATTCGACCAAGTGGGAACGGCAACGGCTCTGAAAGTGAACGCTTCATACAACAACATCATTGTATTGACCGATTCGAACGCACCTAGTATCTTCTCTTCGAATTTTTATCCGCCACCGAAAGCGCAGAGATTGAATTCATATTACGGAGATTTCCCTATATCACCAGGTCTCGGAACTGTGTCCGTAAATTTGAGAGTGTTGGATTTTTCCCACAATAATGTCACATATATCGCGCCTTATTACTTCAGGCACGCCGATTTAACTCTATCCGAGTTATATCTGTCCCACAACAAACTGAGGAACATAACGAGAGAAGTATTCGGTTCTATGGTCATGTTGCAATACCTGGATTTATCCCACAACCACATCTATCACATGGAATACGACTGTTTTAAGAAAGTGAAGAATTTACAGATCATAAACTTGTCCCACAACCACCTCATCGAACTGCCAGTCGGAATGTTCCACGACATGCAATCTCTGACGTCGGTAGACCTCTCAAATAACAACATCAGAAATTTCGCGGAAAATCTGATTATATCACCAGCGTTGGAAAGGTTAGACTTCTCCGATAATGAGCTGTCGAGGGTGCCGACTAATTGCTTAGCCCCAGCCGCGGCTGCCAACTTAGTTGAACTAGATCTGAGCGGGAACACTATACCTACAGTGTCGATGTCCGACTTCGTACAAAGATTCAGGAACCCCGAGCCGGGAGAGTGGCCCGAGGAGTCGGACTACAGCGACGAGTACATGTACCACACGGCTAGACGAGATCACACCAGAGTTTACCACCAAAAAAAGCAATACCCACAAAACGTTTTGTATAAGTCTTTAGCGTGGTTGGATCTGTCGGATAATCATCTGGTACGAATCGAAACGGCTTCGTTCTCGGCGCTGCCCAAGCTGAAGTGGTTGGACATAAGCAACAACAACCCGTTCAATAACGGGGAGAGAGGAAACAATATATTTAAAGGACTCGAAAGACGTCTTACACATTTAGGGTTGAAGAATATACCCTCAATGCTGCTCTCGAAACTGAAGAGTCTGGATCTATCCTACAATAATTTACCATCAGTACCTCCGGATACGACAGCTAACCTAACCCGACTCCGAGTACTGGATTTATCCTACAACGACCTGACAACAGTTCCAGTGGCAACGCATTCACTGACCGAGCTCCGTTGGCTGTCGTTGTCTGGCAACCCCATCACTGCGCTCATGAACACGAGCCTTTACGGAATATCGCCGCGCCTGGAGTACCTGGACGTTACTAGGCTACGTTTGCGAATATTTGAGCTTGGCGCCTTCGGAAAAATGTACGCACTGAGAACTCTCAAAGTGACGTCATATTCGAATCACAGAGACTTCAACATCCCGCGCATGTTGACTTACAACGCCGGACTCGAGAACCTTCTGGTTGAAATCGACGAACTAGGAACCGAATTCGGGAAGGAAATGTACGGAGCACTGCCTTTGAAGCTCAATAATATTACAATTACGGGCCAAAGTTTGAAATACCTAACTCAATATTTGATAAGTGGAGTTCAATCGAGGTCCCTTCGTCTGACGATATACAATACGAGCGTCGAAGAAATAGCCGGCGAAGTATTTTGGCGGCCGGGACGAGTCCGGAACTTGACCTTGGATTTGAGGAATAATTACATTTCTAGAGTACCCAATCCGAGTCAGAGGGGATGGCCCGGGGTTCCGAACTCTTTGTTCCTAGAAGATATATTTGTTGCTGGGAATCCATTGATATGCGACTGCGGCGTCGGATGGATTGAAGCATGGGATAGAAAGAGAAGACAGTATCTTTGTAATGATCCTCACAGTTGTGTGGCGACGAGAGATGACGTCAGATTCGCTACCTGCCCTTCGTATGAAAACAGATCCTTGAGCGATGTACTGTCAAGAGATCTAGATTGTGATTGGAATAAAGGATTTTTAAGTTCACCAAATACGTTCGTAATCATAGGAATGTCAATTCTAACGTTATTATACATTTGA
Protein
MRNNNLVIDLSGERTRSLMAVVKFGYTLVAVTLLLMIWASLSRAIELHVDTGHPPCLFQALCTCSKPAADLGIVTCNHVPLLRVPSTINNSKVFMLQLTGNKIRDLEPQFFQTTGLYRLAINKNPLESIHEEAFYGLDNTLWELELREDKLIAVPSRALRYLQKLRLLDLSGNEITDISGDNWRGLENTLQILILADNSIANLPLDAFSGLPLLETLDLHGNHLSVIDSGVFRDGMSRLNRLLLGDNQLSYIPYEELSPLRQLRVLDLSSNVIKQVPPLHDLNGIKLSLDLLKLDNNMIQVLYPGAFKYFNVLNSTTLDRNPIVTIREDAFRNAKIKSLSLRDCSISDLSPAAFAGLENSLISLDLSENNLTMVSKFMLNKLDSLRFLNLRENKVDISLLTTNMPSEYQQSAFNSFQYRLFSLDVSGANSIEISLQDAKRMRSLRSLSVGKLLRQSIKTEDFLDFGMELEDLKLFGSNLIRIESSAFQYVRTIKTLDLSENNIEYIDPFAFAELHSLITLKMARGLSDSVIILPFEPLKALTELQYLDMSNNKLKNVPDTSFHFLKNLRSLNLEDNAIEHFSKGTLQADIHGKLESICLSLNQMKQIGQHTFVELKEMQEILIEDNLIQYIQRRAFASLDNLKIIKLKGNRINEISEEAFQNIPALKELDVSFNKLETFKFSIFDQVGTATALKVNASYNNIIVLTDSNAPSIFSSNFYPPPKAQRLNSYYGDFPISPGLGTVSVNLRVLDFSHNNVTYIAPYYFRHADLTLSELYLSHNKLRNITREVFGSMVMLQYLDLSHNHIYHMEYDCFKKVKNLQIINLSHNHLIELPVGMFHDMQSLTSVDLSNNNIRNFAENLIISPALERLDFSDNELSRVPTNCLAPAAAANLVELDLSGNTIPTVSMSDFVQRFRNPEPGEWPEESDYSDEYMYHTARRDHTRVYHQKKQYPQNVLYKSLAWLDLSDNHLVRIETASFSALPKLKWLDISNNNPFNNGERGNNIFKGLERRLTHLGLKNIPSMLLSKLKSLDLSYNNLPSVPPDTTANLTRLRVLDLSYNDLTTVPVATHSLTELRWLSLSGNPITALMNTSLYGISPRLEYLDVTRLRLRIFELGAFGKMYALRTLKVTSYSNHRDFNIPRMLTYNAGLENLLVEIDELGTEFGKEMYGALPLKLNNITITGQSLKYLTQYLISGVQSRSLRLTIYNTSVEEIAGEVFWRPGRVRNLTLDLRNNYISRVPNPSQRGWPGVPNSLFLEDIFVAGNPLICDCGVGWIEAWDRKRRQYLCNDPHSCVATRDDVRFATCPSYENRSLSDVLSRDLDCDWNKGFLSSPNTFVIIGMSILTLLYI
Summary
Description
Required for photoreceptor cell morphogenesis. Mediates homophilic cellular adhesion.
Similarity
Belongs to the membrane-bound acyltransferase family.
Belongs to the chaoptin family.
Belongs to the chaoptin family.
Keywords
Cell adhesion
Cell membrane
Complete proteome
Direct protein sequencing
Glycoprotein
Leucine-rich repeat
Membrane
Reference proteome
Repeat
Sensory transduction
Signal
Vision
Feature
chain Chaoptin
Uniprot
H9J9W3
A0A2A4K7F7
A0A194QG22
A0A212EHN3
A0A194RBD0
A0A1W4WDR3
+ More
A0A139WMQ1 A0A232ESX0 K7ITY1 A0A3L8DPM4 A0A088AI45 A0A026WPZ8 A0A2J7PT83 A0A0C9RDW7 A0A1B0FP58 A0A0B4KHK5 A0A2A3EBH2 A0A0B4KHX5 A0A1B6CP54 A0A0B4KHG5 B4PNB0 A0A1I8NSG5 A0A0A1WGJ4 B4NFC1 B4K6G7 F0JAG8 B3M1K3 P12024 E2A0X8 B4M5V1 B4QSK1 A0A158NV56 B3P7S1 B4HZZ9 F4WS44 A0A0B4KI35 A0A182IPR6 A0A1I8NSG2 A0A1I8NSG1 A0A1I8NSG3 A0A1I8NH09 A0A0M5JA11 E2C6Q0 W8AFI2 A0A1W4UI50 B4GN61 A0A195CFH1 A0A034VV46 A0A1I8NSF9 A0A1I8NH07 Q29CE5 A0A3B0JZ76 A0A0L0BY72 A0A1Q3FSG3 A0A0K8V001 B4JYP5 A0A1Q3FT09 A0A084VLQ4 A0A0M9A3U7 B0X0Q6 A0A310SBP2 Q16X81 A0A0J7KTW9 A0A1B6D0B7 F0JAL7 A0A0K8VU17 A0A182XYE2 A0A182R217 A0A182N118 E3CTQ1 E3CTS9 A0A182PNT2 A0A154PE71 A0A182M164 A0A0K8WJJ8 Q5TUS9 B7FNR3 A0A1S4G8C7 A0A1B0AJW3 A0A182QLG2 A0A182XNN1 A0A182VBH2 A0A182HIU6 A0A182UBZ4 A0A195F4E1 A0A1B6F7H5 A0A0T6AZL7 A0A151XE86 A0A195DZQ7 A0A2S2QXL6 J9JN12 E9JCB6 A0A2H8TLK4 A0A182VVJ1 E0VD25 A0A0P6BPZ4 A0A0P6HGB5 A0A0P5ZFV8
A0A139WMQ1 A0A232ESX0 K7ITY1 A0A3L8DPM4 A0A088AI45 A0A026WPZ8 A0A2J7PT83 A0A0C9RDW7 A0A1B0FP58 A0A0B4KHK5 A0A2A3EBH2 A0A0B4KHX5 A0A1B6CP54 A0A0B4KHG5 B4PNB0 A0A1I8NSG5 A0A0A1WGJ4 B4NFC1 B4K6G7 F0JAG8 B3M1K3 P12024 E2A0X8 B4M5V1 B4QSK1 A0A158NV56 B3P7S1 B4HZZ9 F4WS44 A0A0B4KI35 A0A182IPR6 A0A1I8NSG2 A0A1I8NSG1 A0A1I8NSG3 A0A1I8NH09 A0A0M5JA11 E2C6Q0 W8AFI2 A0A1W4UI50 B4GN61 A0A195CFH1 A0A034VV46 A0A1I8NSF9 A0A1I8NH07 Q29CE5 A0A3B0JZ76 A0A0L0BY72 A0A1Q3FSG3 A0A0K8V001 B4JYP5 A0A1Q3FT09 A0A084VLQ4 A0A0M9A3U7 B0X0Q6 A0A310SBP2 Q16X81 A0A0J7KTW9 A0A1B6D0B7 F0JAL7 A0A0K8VU17 A0A182XYE2 A0A182R217 A0A182N118 E3CTQ1 E3CTS9 A0A182PNT2 A0A154PE71 A0A182M164 A0A0K8WJJ8 Q5TUS9 B7FNR3 A0A1S4G8C7 A0A1B0AJW3 A0A182QLG2 A0A182XNN1 A0A182VBH2 A0A182HIU6 A0A182UBZ4 A0A195F4E1 A0A1B6F7H5 A0A0T6AZL7 A0A151XE86 A0A195DZQ7 A0A2S2QXL6 J9JN12 E9JCB6 A0A2H8TLK4 A0A182VVJ1 E0VD25 A0A0P6BPZ4 A0A0P6HGB5 A0A0P5ZFV8
Pubmed
19121390
26354079
22118469
18362917
19820115
28648823
+ More
20075255 30249741 24508170 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17994087 17550304 25830018 3124963 3920657 6420071 17893096 19415110 20798317 21347285 21719571 25315136 24495485 25348373 15632085 26108605 24438588 17510324 25244985 12364791 21282665 20566863
20075255 30249741 24508170 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17994087 17550304 25830018 3124963 3920657 6420071 17893096 19415110 20798317 21347285 21719571 25315136 24495485 25348373 15632085 26108605 24438588 17510324 25244985 12364791 21282665 20566863
EMBL
BABH01025981
BABH01025982
BABH01025983
BABH01025984
NWSH01000077
PCG79868.1
+ More
KQ459249 KPJ02396.1 AGBW02014861 OWR40981.1 KQ460647 KPJ13206.1 KQ971312 KYB29164.1 NNAY01002364 OXU21458.1 QOIP01000006 RLU22192.1 KK107135 EZA58102.1 NEVH01021919 PNF19535.1 GBYB01011277 JAG81044.1 CCAG010007645 AE014297 AGB96514.1 KZ288293 PBC29040.1 AGB96516.1 GEDC01022147 JAS15151.1 AGB96515.1 CM000160 EDW99192.2 GBXI01016290 GBXI01011563 JAC98001.1 JAD02729.1 CH964251 EDW82988.2 CH933806 EDW16267.2 BT126010 ADY17709.1 CH902617 EDV43294.2 M19017 M19008 M19009 M19010 M19011 M19012 M19013 M19014 M19016 K03274 GL435626 EFN73031.1 CH940652 EDW59027.2 CM000364 EDX15098.1 ADTU01026899 CH954182 EDV52979.1 CH480819 EDW53606.1 GL888296 EGI62972.1 AGB96513.1 CP012526 ALC48036.1 GL453161 EFN76362.1 GAMC01019575 JAB86980.1 CH479186 EDW39187.1 KQ977827 KYM99465.1 GAKP01012638 JAC46314.1 CM000070 EAL26701.2 OUUW01000005 SPP80820.1 JRES01001160 KNC24983.1 GFDL01004528 JAV30517.1 GDHF01020100 JAI32214.1 CH916377 EDV90807.1 GFDL01004492 JAV30553.1 ATLV01014543 KE524974 KFB38898.1 KQ435742 KOX76667.1 DS232243 EDS38299.1 KQ770716 OAD52623.1 CH477545 EAT39239.1 LBMM01003239 KMQ93768.1 GEDC01018129 JAS19169.1 BT126059 ADY17758.1 GDHF01028643 GDHF01009937 JAI23671.1 JAI42377.1 BT125717 ADO51079.1 BT125745 ADP38085.1 KQ434886 KZC10143.1 AXCM01009692 GDHF01000981 JAI51333.1 AAAB01008846 EAL41313.4 BT053753 ACK77671.1 AXCN02001386 APCN01004099 KQ981820 KYN35251.1 GECZ01023644 JAS46125.1 LJIG01022445 KRT80569.1 KQ982254 KYQ58673.1 KQ980031 KYN18187.1 GGMS01013316 MBY82519.1 ABLF02035208 ABLF02035212 ABLF02035218 GL771786 EFZ09539.1 GFXV01003114 MBW14919.1 DS235070 EEB11281.1 GDIP01026596 JAM77119.1 GDIQ01019867 JAN74870.1 GDIP01044392 JAM59323.1
KQ459249 KPJ02396.1 AGBW02014861 OWR40981.1 KQ460647 KPJ13206.1 KQ971312 KYB29164.1 NNAY01002364 OXU21458.1 QOIP01000006 RLU22192.1 KK107135 EZA58102.1 NEVH01021919 PNF19535.1 GBYB01011277 JAG81044.1 CCAG010007645 AE014297 AGB96514.1 KZ288293 PBC29040.1 AGB96516.1 GEDC01022147 JAS15151.1 AGB96515.1 CM000160 EDW99192.2 GBXI01016290 GBXI01011563 JAC98001.1 JAD02729.1 CH964251 EDW82988.2 CH933806 EDW16267.2 BT126010 ADY17709.1 CH902617 EDV43294.2 M19017 M19008 M19009 M19010 M19011 M19012 M19013 M19014 M19016 K03274 GL435626 EFN73031.1 CH940652 EDW59027.2 CM000364 EDX15098.1 ADTU01026899 CH954182 EDV52979.1 CH480819 EDW53606.1 GL888296 EGI62972.1 AGB96513.1 CP012526 ALC48036.1 GL453161 EFN76362.1 GAMC01019575 JAB86980.1 CH479186 EDW39187.1 KQ977827 KYM99465.1 GAKP01012638 JAC46314.1 CM000070 EAL26701.2 OUUW01000005 SPP80820.1 JRES01001160 KNC24983.1 GFDL01004528 JAV30517.1 GDHF01020100 JAI32214.1 CH916377 EDV90807.1 GFDL01004492 JAV30553.1 ATLV01014543 KE524974 KFB38898.1 KQ435742 KOX76667.1 DS232243 EDS38299.1 KQ770716 OAD52623.1 CH477545 EAT39239.1 LBMM01003239 KMQ93768.1 GEDC01018129 JAS19169.1 BT126059 ADY17758.1 GDHF01028643 GDHF01009937 JAI23671.1 JAI42377.1 BT125717 ADO51079.1 BT125745 ADP38085.1 KQ434886 KZC10143.1 AXCM01009692 GDHF01000981 JAI51333.1 AAAB01008846 EAL41313.4 BT053753 ACK77671.1 AXCN02001386 APCN01004099 KQ981820 KYN35251.1 GECZ01023644 JAS46125.1 LJIG01022445 KRT80569.1 KQ982254 KYQ58673.1 KQ980031 KYN18187.1 GGMS01013316 MBY82519.1 ABLF02035208 ABLF02035212 ABLF02035218 GL771786 EFZ09539.1 GFXV01003114 MBW14919.1 DS235070 EEB11281.1 GDIP01026596 JAM77119.1 GDIQ01019867 JAN74870.1 GDIP01044392 JAM59323.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000053240
UP000192223
+ More
UP000007266 UP000215335 UP000002358 UP000279307 UP000005203 UP000053097 UP000235965 UP000092444 UP000000803 UP000242457 UP000002282 UP000095300 UP000007798 UP000009192 UP000007801 UP000000311 UP000008792 UP000000304 UP000005205 UP000008711 UP000001292 UP000007755 UP000075880 UP000095301 UP000092553 UP000008237 UP000192221 UP000008744 UP000078542 UP000001819 UP000268350 UP000037069 UP000001070 UP000030765 UP000053105 UP000002320 UP000008820 UP000036403 UP000076408 UP000075900 UP000075884 UP000075885 UP000076502 UP000075883 UP000007062 UP000092445 UP000075886 UP000076407 UP000075903 UP000075840 UP000075902 UP000078541 UP000075809 UP000078492 UP000007819 UP000075920 UP000009046
UP000007266 UP000215335 UP000002358 UP000279307 UP000005203 UP000053097 UP000235965 UP000092444 UP000000803 UP000242457 UP000002282 UP000095300 UP000007798 UP000009192 UP000007801 UP000000311 UP000008792 UP000000304 UP000005205 UP000008711 UP000001292 UP000007755 UP000075880 UP000095301 UP000092553 UP000008237 UP000192221 UP000008744 UP000078542 UP000001819 UP000268350 UP000037069 UP000001070 UP000030765 UP000053105 UP000002320 UP000008820 UP000036403 UP000076408 UP000075900 UP000075884 UP000075885 UP000076502 UP000075883 UP000007062 UP000092445 UP000075886 UP000076407 UP000075903 UP000075840 UP000075902 UP000078541 UP000075809 UP000078492 UP000007819 UP000075920 UP000009046
Pfam
Interpro
Gene 3D
ProteinModelPortal
H9J9W3
A0A2A4K7F7
A0A194QG22
A0A212EHN3
A0A194RBD0
A0A1W4WDR3
+ More
A0A139WMQ1 A0A232ESX0 K7ITY1 A0A3L8DPM4 A0A088AI45 A0A026WPZ8 A0A2J7PT83 A0A0C9RDW7 A0A1B0FP58 A0A0B4KHK5 A0A2A3EBH2 A0A0B4KHX5 A0A1B6CP54 A0A0B4KHG5 B4PNB0 A0A1I8NSG5 A0A0A1WGJ4 B4NFC1 B4K6G7 F0JAG8 B3M1K3 P12024 E2A0X8 B4M5V1 B4QSK1 A0A158NV56 B3P7S1 B4HZZ9 F4WS44 A0A0B4KI35 A0A182IPR6 A0A1I8NSG2 A0A1I8NSG1 A0A1I8NSG3 A0A1I8NH09 A0A0M5JA11 E2C6Q0 W8AFI2 A0A1W4UI50 B4GN61 A0A195CFH1 A0A034VV46 A0A1I8NSF9 A0A1I8NH07 Q29CE5 A0A3B0JZ76 A0A0L0BY72 A0A1Q3FSG3 A0A0K8V001 B4JYP5 A0A1Q3FT09 A0A084VLQ4 A0A0M9A3U7 B0X0Q6 A0A310SBP2 Q16X81 A0A0J7KTW9 A0A1B6D0B7 F0JAL7 A0A0K8VU17 A0A182XYE2 A0A182R217 A0A182N118 E3CTQ1 E3CTS9 A0A182PNT2 A0A154PE71 A0A182M164 A0A0K8WJJ8 Q5TUS9 B7FNR3 A0A1S4G8C7 A0A1B0AJW3 A0A182QLG2 A0A182XNN1 A0A182VBH2 A0A182HIU6 A0A182UBZ4 A0A195F4E1 A0A1B6F7H5 A0A0T6AZL7 A0A151XE86 A0A195DZQ7 A0A2S2QXL6 J9JN12 E9JCB6 A0A2H8TLK4 A0A182VVJ1 E0VD25 A0A0P6BPZ4 A0A0P6HGB5 A0A0P5ZFV8
A0A139WMQ1 A0A232ESX0 K7ITY1 A0A3L8DPM4 A0A088AI45 A0A026WPZ8 A0A2J7PT83 A0A0C9RDW7 A0A1B0FP58 A0A0B4KHK5 A0A2A3EBH2 A0A0B4KHX5 A0A1B6CP54 A0A0B4KHG5 B4PNB0 A0A1I8NSG5 A0A0A1WGJ4 B4NFC1 B4K6G7 F0JAG8 B3M1K3 P12024 E2A0X8 B4M5V1 B4QSK1 A0A158NV56 B3P7S1 B4HZZ9 F4WS44 A0A0B4KI35 A0A182IPR6 A0A1I8NSG2 A0A1I8NSG1 A0A1I8NSG3 A0A1I8NH09 A0A0M5JA11 E2C6Q0 W8AFI2 A0A1W4UI50 B4GN61 A0A195CFH1 A0A034VV46 A0A1I8NSF9 A0A1I8NH07 Q29CE5 A0A3B0JZ76 A0A0L0BY72 A0A1Q3FSG3 A0A0K8V001 B4JYP5 A0A1Q3FT09 A0A084VLQ4 A0A0M9A3U7 B0X0Q6 A0A310SBP2 Q16X81 A0A0J7KTW9 A0A1B6D0B7 F0JAL7 A0A0K8VU17 A0A182XYE2 A0A182R217 A0A182N118 E3CTQ1 E3CTS9 A0A182PNT2 A0A154PE71 A0A182M164 A0A0K8WJJ8 Q5TUS9 B7FNR3 A0A1S4G8C7 A0A1B0AJW3 A0A182QLG2 A0A182XNN1 A0A182VBH2 A0A182HIU6 A0A182UBZ4 A0A195F4E1 A0A1B6F7H5 A0A0T6AZL7 A0A151XE86 A0A195DZQ7 A0A2S2QXL6 J9JN12 E9JCB6 A0A2H8TLK4 A0A182VVJ1 E0VD25 A0A0P6BPZ4 A0A0P6HGB5 A0A0P5ZFV8
PDB
4KNG
E-value=2.32196e-23,
Score=275
Ontologies
GO
Topology
Subcellular location
Cell membrane
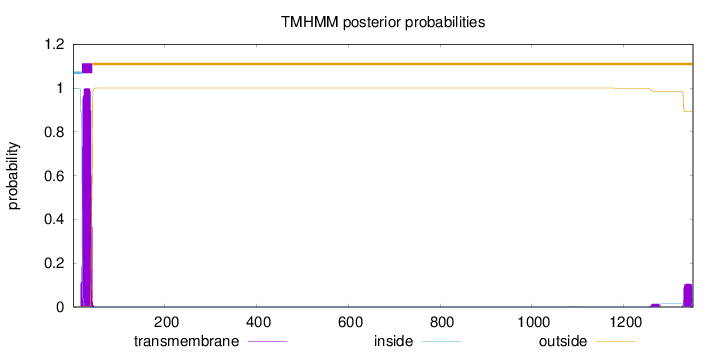
Length:
1351
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
23.76643
Exp number, first 60 AAs:
21.38636
Total prob of N-in:
0.99826
POSSIBLE N-term signal
sequence
inside
1 - 19
TMhelix
20 - 42
outside
43 - 1351
Population Genetic Test Statistics
Pi
244.794704
Theta
170.91046
Tajima's D
1.47912
CLR
0.3217
CSRT
0.778561071946403
Interpretation
Uncertain
