Gene
KWMTBOMO03573 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA006282
Annotation
PREDICTED:_uncharacterized_protein_LOC106120359_[Papilio_xuthus]
Location in the cell
Lysosomal Reliability : 1.266
Sequence
CDS
ATGAAGGCTTTGGCAGCTGTACTATTGTTAGTTGTGGCAGTGAACGGTGCGTGGAAAGGTGGACTGCGGACAACATGGGGTGTAGGCCTTGGTCTAGGAACGGATTTTTTCTATGAGATTCCAAGGTCACTGGAAGAGGTTAAGTCTCAGGGCTGGGTTTTAACCGACAAAGCTGACGGACTTCCTCTTCCTTCGCTCGCCTTGTATTGTTCTGAAGACCGCATCTTATGCGGATTTTTTGACGAAACCGAATATGTGGTGGGATTGCAGGTTTCTTTACCCGTAGATGAGGTCACGGACATTATTTTTGATATGCCAACCCAAGGTTTTACAATATGGGAAACAGAAGTGGACGGGAAACTTAAAAAATTCTACAGTATCCAACAATACTTCATCAATGAAGATACTCTAAAACAAAGTGTTGAGGATCGTTTGAGTAAGTGGGATAGTTCCAAGACTCTTCAAGAAAAATCCATTTGGGTAACCGGCTTCAATGGTACCCTTCTTGAAATATCCACCGTTGCTGATGATATTGCTAACGGAGATGATTTCACTAAGCAAGCCTGTGTTCCTTGGATGGGGCGTCATTATTACTACAAGATGTCTGCGGAGACGGAGTGCAAAGCTGACACATTGTTACCATGGTTTCCGATTGTTGAATCCGGGGAATTGATCGCTACCGGTTTTATTAGCTTCCTGAAACTTTCGAACACTTACAAATGGTTTGAGAAGCCTTCGAAAGCAGCTGTTCAATTCATTGTTCCTGATGGCCCTAAGTGCTTATACGATTTGGCTGAGGATTCTGGACTAACAACAATGCACATCTATTATGTCAATCAACCATGGTTGGTCAGTTGCCTGTGGCAGTGA
Protein
MKALAAVLLLVVAVNGAWKGGLRTTWGVGLGLGTDFFYEIPRSLEEVKSQGWVLTDKADGLPLPSLALYCSEDRILCGFFDETEYVVGLQVSLPVDEVTDIIFDMPTQGFTIWETEVDGKLKKFYSIQQYFINEDTLKQSVEDRLSKWDSSKTLQEKSIWVTGFNGTLLEISTVADDIANGDDFTKQACVPWMGRHYYYKMSAETECKADTLLPWFPIVESGELIATGFISFLKLSNTYKWFEKPSKAAVQFIVPDGPKCLYDLAEDSGLTTMHIYYVNQPWLVSCLWQ
Summary
Uniprot
A0A348ARE3
H9J9T8
A0A194Q7G6
A0A194RJL1
A0A194RPR4
A0A212F8X0
+ More
A0A194RKF0 A0A2A4J3W2 A0A2W1BGD7 A0A2A4J3P8 A0A2A4J2K5 A0A2W1B0B8 A0A212F8Y2 A0A2W1B5C2 A0A2A4J3S8 A0A2H1X436 B1NLD7 A0A2H1WZU4 A0A2H1WZI2 A0A2K8JLE3 A0A0A9XW04 A0A146L760 A0A0A9WTE6 A0A1D1ZDW1 A0A1B6JCM2 A0A1B6L089 A0A1B6KUQ9 A0A1B6M7K5 A0A1B6F1Y6 A0A3G9CLZ2 A0A2R7W9Q1 C3Y5T8 A0A210R159 A0A1B6KHA3 A0A1B6C8E0 C3Y5U1 T1IXM1 A0A1S3JDE4 A0A1B6IDP6 V4C0Z0 A0A226F3Q4 A0A1B6IFE9 A0A1W7R968 T1J4X8 A0A1D2NIT9 V4ADY4 A0A1I8GH03 A0A1I8HZZ2 A0A1D2N6X1 A0A1I8ITE8 A0A226E1C8 K1QTT1 A0A210R5Y3 A0A0K2TFV4 C3Y5U4 A0A0B6YRW2 A0A2C9KD43 C3Y5U2 C3Y5U3 A0A226ERP2 A0A0K2VK12 T1ISJ9 T1JA23 A0A1B6HGZ3 A0A0K2VJM8 A0A1B6LB36 A0A1B6JIB1 A0A1B6I8C4 A0A1S3IAF0 A0A2B4SJL6 A0A226DNZ2
A0A194RKF0 A0A2A4J3W2 A0A2W1BGD7 A0A2A4J3P8 A0A2A4J2K5 A0A2W1B0B8 A0A212F8Y2 A0A2W1B5C2 A0A2A4J3S8 A0A2H1X436 B1NLD7 A0A2H1WZU4 A0A2H1WZI2 A0A2K8JLE3 A0A0A9XW04 A0A146L760 A0A0A9WTE6 A0A1D1ZDW1 A0A1B6JCM2 A0A1B6L089 A0A1B6KUQ9 A0A1B6M7K5 A0A1B6F1Y6 A0A3G9CLZ2 A0A2R7W9Q1 C3Y5T8 A0A210R159 A0A1B6KHA3 A0A1B6C8E0 C3Y5U1 T1IXM1 A0A1S3JDE4 A0A1B6IDP6 V4C0Z0 A0A226F3Q4 A0A1B6IFE9 A0A1W7R968 T1J4X8 A0A1D2NIT9 V4ADY4 A0A1I8GH03 A0A1I8HZZ2 A0A1D2N6X1 A0A1I8ITE8 A0A226E1C8 K1QTT1 A0A210R5Y3 A0A0K2TFV4 C3Y5U4 A0A0B6YRW2 A0A2C9KD43 C3Y5U2 C3Y5U3 A0A226ERP2 A0A0K2VK12 T1ISJ9 T1JA23 A0A1B6HGZ3 A0A0K2VJM8 A0A1B6LB36 A0A1B6JIB1 A0A1B6I8C4 A0A1S3IAF0 A0A2B4SJL6 A0A226DNZ2
Pubmed
EMBL
LC259005
BBC08867.1
BABH01025962
KQ459324
KPJ01482.1
KQ460045
+ More
KPJ18018.1 KPJ18016.1 AGBW02009658 OWR50182.1 KPJ18017.1 NWSH01003255 PCG66681.1 KZ150247 PZC71860.1 PCG66685.1 NWSH01003566 PCG66099.1 KZ150517 PZC70622.1 PZC71857.1 OWR50181.1 PZC70621.1 PCG66100.1 ODYU01013295 SOQ60002.1 EF600052 ABU98617.1 ODYU01012270 SOQ58486.1 SOQ58485.1 MF683358 ATU82499.1 GBHO01018617 JAG24987.1 GDHC01015627 JAQ03002.1 GBHO01033826 JAG09778.1 GDJX01002923 JAT65013.1 GECU01010695 JAS97011.1 GEBQ01022916 JAT17061.1 GEBQ01024993 JAT14984.1 GEBQ01008045 JAT31932.1 GECZ01025473 JAS44296.1 FX983003 BAS30471.1 KK854485 PTY16269.1 GG666487 EEN64335.1 NEDP02000926 OWF54729.1 GEBQ01029151 JAT10826.1 GEDC01027798 JAS09500.1 EEN64338.1 AFFK01020108 GECU01022662 GECU01019782 JAS85044.1 JAS87924.1 KB201701 ESO95124.1 LNIX01000001 OXA63990.1 GECU01022084 JAS85622.1 GFAH01000703 JAV47686.1 JH431850 LJIJ01000029 ODN05174.1 KB201931 ESO93325.1 NIVC01002056 PAA61235.1 NIVC01002351 PAA58643.1 LJIJ01000185 ODN00825.1 LNIX01000008 OXA51100.1 JH816663 EKC34624.1 NEDP02000192 OWF56459.1 HACA01007542 CDW24903.1 EEN64341.1 HACG01012134 CEK58999.1 EEN64339.1 EEN64340.1 LNIX01000002 OXA59724.1 HACA01033304 CDW50665.1 JH431434 JH431981 GECU01033736 JAS73970.1 HACA01033303 CDW50664.1 GEBQ01019079 JAT20898.1 GECU01008864 JAS98842.1 GECU01024566 JAS83140.1 LSMT01000044 PFX30874.1 LNIX01000013 OXA47255.1
KPJ18018.1 KPJ18016.1 AGBW02009658 OWR50182.1 KPJ18017.1 NWSH01003255 PCG66681.1 KZ150247 PZC71860.1 PCG66685.1 NWSH01003566 PCG66099.1 KZ150517 PZC70622.1 PZC71857.1 OWR50181.1 PZC70621.1 PCG66100.1 ODYU01013295 SOQ60002.1 EF600052 ABU98617.1 ODYU01012270 SOQ58486.1 SOQ58485.1 MF683358 ATU82499.1 GBHO01018617 JAG24987.1 GDHC01015627 JAQ03002.1 GBHO01033826 JAG09778.1 GDJX01002923 JAT65013.1 GECU01010695 JAS97011.1 GEBQ01022916 JAT17061.1 GEBQ01024993 JAT14984.1 GEBQ01008045 JAT31932.1 GECZ01025473 JAS44296.1 FX983003 BAS30471.1 KK854485 PTY16269.1 GG666487 EEN64335.1 NEDP02000926 OWF54729.1 GEBQ01029151 JAT10826.1 GEDC01027798 JAS09500.1 EEN64338.1 AFFK01020108 GECU01022662 GECU01019782 JAS85044.1 JAS87924.1 KB201701 ESO95124.1 LNIX01000001 OXA63990.1 GECU01022084 JAS85622.1 GFAH01000703 JAV47686.1 JH431850 LJIJ01000029 ODN05174.1 KB201931 ESO93325.1 NIVC01002056 PAA61235.1 NIVC01002351 PAA58643.1 LJIJ01000185 ODN00825.1 LNIX01000008 OXA51100.1 JH816663 EKC34624.1 NEDP02000192 OWF56459.1 HACA01007542 CDW24903.1 EEN64341.1 HACG01012134 CEK58999.1 EEN64339.1 EEN64340.1 LNIX01000002 OXA59724.1 HACA01033304 CDW50665.1 JH431434 JH431981 GECU01033736 JAS73970.1 HACA01033303 CDW50664.1 GEBQ01019079 JAT20898.1 GECU01008864 JAS98842.1 GECU01024566 JAS83140.1 LSMT01000044 PFX30874.1 LNIX01000013 OXA47255.1
Proteomes
PRIDE
Pfam
Interpro
Gene 3D
ProteinModelPortal
A0A348ARE3
H9J9T8
A0A194Q7G6
A0A194RJL1
A0A194RPR4
A0A212F8X0
+ More
A0A194RKF0 A0A2A4J3W2 A0A2W1BGD7 A0A2A4J3P8 A0A2A4J2K5 A0A2W1B0B8 A0A212F8Y2 A0A2W1B5C2 A0A2A4J3S8 A0A2H1X436 B1NLD7 A0A2H1WZU4 A0A2H1WZI2 A0A2K8JLE3 A0A0A9XW04 A0A146L760 A0A0A9WTE6 A0A1D1ZDW1 A0A1B6JCM2 A0A1B6L089 A0A1B6KUQ9 A0A1B6M7K5 A0A1B6F1Y6 A0A3G9CLZ2 A0A2R7W9Q1 C3Y5T8 A0A210R159 A0A1B6KHA3 A0A1B6C8E0 C3Y5U1 T1IXM1 A0A1S3JDE4 A0A1B6IDP6 V4C0Z0 A0A226F3Q4 A0A1B6IFE9 A0A1W7R968 T1J4X8 A0A1D2NIT9 V4ADY4 A0A1I8GH03 A0A1I8HZZ2 A0A1D2N6X1 A0A1I8ITE8 A0A226E1C8 K1QTT1 A0A210R5Y3 A0A0K2TFV4 C3Y5U4 A0A0B6YRW2 A0A2C9KD43 C3Y5U2 C3Y5U3 A0A226ERP2 A0A0K2VK12 T1ISJ9 T1JA23 A0A1B6HGZ3 A0A0K2VJM8 A0A1B6LB36 A0A1B6JIB1 A0A1B6I8C4 A0A1S3IAF0 A0A2B4SJL6 A0A226DNZ2
A0A194RKF0 A0A2A4J3W2 A0A2W1BGD7 A0A2A4J3P8 A0A2A4J2K5 A0A2W1B0B8 A0A212F8Y2 A0A2W1B5C2 A0A2A4J3S8 A0A2H1X436 B1NLD7 A0A2H1WZU4 A0A2H1WZI2 A0A2K8JLE3 A0A0A9XW04 A0A146L760 A0A0A9WTE6 A0A1D1ZDW1 A0A1B6JCM2 A0A1B6L089 A0A1B6KUQ9 A0A1B6M7K5 A0A1B6F1Y6 A0A3G9CLZ2 A0A2R7W9Q1 C3Y5T8 A0A210R159 A0A1B6KHA3 A0A1B6C8E0 C3Y5U1 T1IXM1 A0A1S3JDE4 A0A1B6IDP6 V4C0Z0 A0A226F3Q4 A0A1B6IFE9 A0A1W7R968 T1J4X8 A0A1D2NIT9 V4ADY4 A0A1I8GH03 A0A1I8HZZ2 A0A1D2N6X1 A0A1I8ITE8 A0A226E1C8 K1QTT1 A0A210R5Y3 A0A0K2TFV4 C3Y5U4 A0A0B6YRW2 A0A2C9KD43 C3Y5U2 C3Y5U3 A0A226ERP2 A0A0K2VK12 T1ISJ9 T1JA23 A0A1B6HGZ3 A0A0K2VJM8 A0A1B6LB36 A0A1B6JIB1 A0A1B6I8C4 A0A1S3IAF0 A0A2B4SJL6 A0A226DNZ2
Ontologies
Topology
SignalP
Position: 1 - 16,
Likelihood: 0.950551
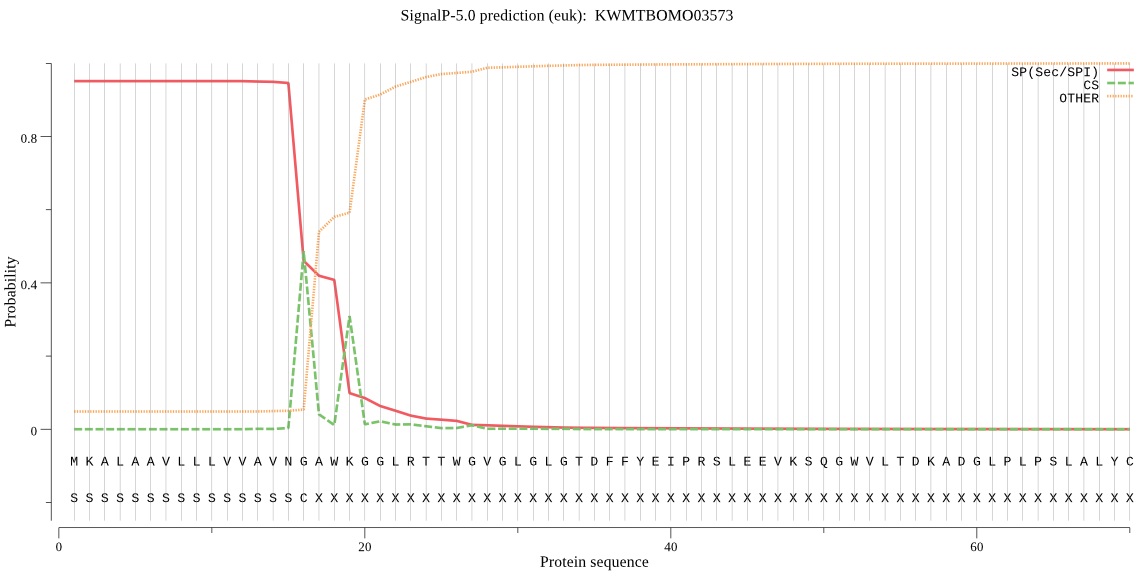
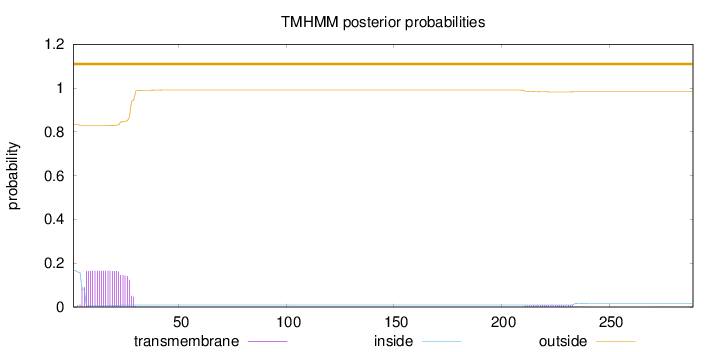
Length:
289
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
3.84983
Exp number, first 60 AAs:
3.64549
Total prob of N-in:
0.16711
outside
1 - 289
Population Genetic Test Statistics
Pi
158.848229
Theta
138.368061
Tajima's D
-1.032289
CLR
0.824236
CSRT
0.131393430328484
Interpretation
Uncertain
