Gene
KWMTBOMO03560
Pre Gene Modal
BGIBMGA006298
Annotation
PREDICTED:_peripheral_plasma_membrane_protein_CASK_[Bombyx_mori]
Full name
Peripheral plasma membrane protein CASK
Alternative Name
Calcium/calmodulin-dependent protein kinase
Location in the cell
Cytoplasmic Reliability : 2.08
Sequence
CDS
ATGGCCGAAGACGAAGTGTTATTCGACGATGTGTACGAATTGTGCGAAATCATCGGAAAGGGTCCATTCAGTTTGGTTCGTCGCTGTGTGCACCGGCAAACAGCACAACAGTTTGCAGTGAAAATTGTGGATGTGGCCCGCTTCACTGCTAGTCCGGGACTTAGCACTGCAGATTTGAAAAGAGAAGCAACAATATGTCATATGTTGAAGCATCCTCATATAGTGGAACTGCTGGAAACCTACAGCTCCGAAGGCATGCTATATATGGTGTTTGAATATATGGATGGCTCTGACCTTTGCTTCGAAGTGGTTCGACGAGCTTCTGCCGGGTTCGTCTACAGCGAAGCGGTGGCATGTCATTACATGCGTCAGATTCTCGAAGCGCTCCGCTATTGTCACGAGAACGACATCGTGCACCGTGACGTCAGACCTCACTGCGTTCTGCTGGCCGGGAGGGACAACTGCGCGCCGGTGAAGCTCGGGGGATTCGGCGTGGCCGCCCAGTTGCCAACGCCCACGTCGCATCGAGCCCCACCAGAACTGGTGATGGACAATCGACCGCTAATGGGTCCGATGGGCCAAGCCTATCTCAAATCCGATGAGTACGGTCGTATCGGTACTCCTCACTATATGGCCCCGGAAGTTGTATCCAGCCAGTTGTACGGGAAGCCCGTCGATGTGTGGGCGGCGGGTATATTGTTGCACGTACTGCTTGTTGGATATCTACCCTTCGCCGGAACAAGGGAACGCCTGTTCGAGGCTATTTGCAGGGGGAGATTAAGGTTCGACGCTCCTCTCTGGGATGATGTAAGCGATGCTGCCAAAGATCTAGTGCAACGCATGCTCACCGTCGATCACACTCAGAGGATCAACATTCAAGACGTGCTCAATCATCGATGGATAAAAGATCGCGAAAAACAGAACCGCATCCACCTGGCCGGTACCGTCGAGGAGCTGAAGAAGTTTAATAGTCGTCGTCGATTGAGGGCTGCCACTCTCGGCGCCGCCGCGGCCGACGACGAGGAGGAGGAGGAACAGCAGCCGACTCGCAAACAGGCGATGCTCGAGGAGGCCAACGCTGCAGCTGTGAATATCGTCGTGGAGTCATTGGACGACGTGGCTGTCCTCCAGGACGGTCCGTTGTTCATAGACCCGGACCTCTTCCACAGCGTGCTCGACGATTTGCAACTGAGAGCCCTACTAGAGGTGTACGACCGCATAGCGTGCACTGTGGTGGTGACGAGCGGGCGAGCCCCCGCGGCGGAGGGTCGCGCCCGCGCCCGCCACGCGCTGGAGGCCACCACGCGCCTGCGCCACGCCGCCGCCCCCGCCGCGCCCCCCGCCGCGCCCGCCGCCGCCGCGCCCGCCGCCGCCGCGCCCGCCACCGCCGCCGCCGTGCACGACCTGCGCCGCCTGCTCGCTCTGCCGCACGTTAAGAAAATCTAA
Protein
MAEDEVLFDDVYELCEIIGKGPFSLVRRCVHRQTAQQFAVKIVDVARFTASPGLSTADLKREATICHMLKHPHIVELLETYSSEGMLYMVFEYMDGSDLCFEVVRRASAGFVYSEAVACHYMRQILEALRYCHENDIVHRDVRPHCVLLAGRDNCAPVKLGGFGVAAQLPTPTSHRAPPELVMDNRPLMGPMGQAYLKSDEYGRIGTPHYMAPEVVSSQLYGKPVDVWAAGILLHVLLVGYLPFAGTRERLFEAICRGRLRFDAPLWDDVSDAAKDLVQRMLTVDHTQRINIQDVLNHRWIKDREKQNRIHLAGTVEELKKFNSRRRLRAATLGAAAADDEEEEEQQPTRKQAMLEEANAAAVNIVVESLDDVAVLQDGPLFIDPDLFHSVLDDLQLRALLEVYDRIACTVVVTSGRAPAAEGRARARHALEATTRLRHAAAPAAPPAAPAAAAPAAAAPATAAAVHDLRRLLALPHVKKI
Summary
Description
May regulate transmembrane proteins that bind calcium, calmodulin, or nucleotides. Functionally modulates eag potassium channels; increases eag current and whole-cell conductance. Also regulates autophosphorylation of CaMKII.
Catalytic Activity
ATP + L-seryl-[protein] = ADP + H(+) + O-phospho-L-seryl-[protein]
ATP + L-threonyl-[protein] = ADP + H(+) + O-phospho-L-threonyl-[protein]
ATP + L-threonyl-[protein] = ADP + H(+) + O-phospho-L-threonyl-[protein]
Subunit
Interacts with eag. Interacts with CaMKII.
Similarity
Belongs to the MAGUK family.
In the N-terminal section; belongs to the protein kinase superfamily. CAMK Ser/Thr protein kinase family. CaMK subfamily.
In the N-terminal section; belongs to the protein kinase superfamily. CAMK Ser/Thr protein kinase family. CaMK subfamily.
Keywords
Alternative splicing
ATP-binding
Calmodulin-binding
Cell membrane
Complete proteome
Kinase
Membrane
Nucleotide-binding
Phosphoprotein
Reference proteome
Repeat
SH3 domain
Transferase
Feature
chain Peripheral plasma membrane protein CASK
splice variant In isoform A and isoform G.
splice variant In isoform A and isoform G.
Uniprot
A0A0N0PD55
A0A212F0U0
H9J9V3
A0A194QG32
A0A2A4JFP8
A0A2A4JGK0
+ More
A0A1B6C0X7 A0A1B6DHA6 A0A224XLD3 A0A0K8SEW2 A0A1B6DER4 A0A232F5Q3 A0A1B6G1C8 A0A0N1IT67 A0A0A9WSH7 A0A023F4C5 A0A067RCY7 A0A0A9WSE7 A0A0K8SEV3 A0A0A9WH75 A0A1S4ENM9 A0A2A3EEW1 K7J2A6 K7J2A7 E2ASJ0 A0A2S2Q3F2 E2BZU4 A0A2H8TJY2 A0A0A9WK55 A0A026X1H1 A0A2M4AVN7 A0A0L7RF97 B3P8S1 F5HMI9 B4G619 J9JVX3 B4QZ94 A0A139WKH6 D6WEC4 A0A0Q9VZW4 A0A1A9YUC0 B4JGQ4 A0A0K8U4D6 A0A0K8USR1 A0A0Q9WC59 A0A0Q9VZV6 B4HM48 W8BF57 A0A1A9W5Y9 A0A2C9GQA3 A0A084VVT9 A0A2M4CSE2 A0NGZ6 B3LVH5 A0A0Q9XA77 A0A1W4W1R1 A0A0Q9X046 Q24210 A0A1W4VQ70 Q29AR4 A0A0P8XYE7 A0A3B0JPX6 A0A0P9C8I4 E1JIS7 B4MC57 A0A0R1E448 A0A1W4W370 A0A1W4VQG9 A0A1I8NGR6 A0A0B4KH39 A0A034VQY7 A0A0K8WM54 A0A1I8NGR2 A0A0K8WLW8 A0A0A1X353 A0A0K8VA90 A0A1Y1M581 A0A0K8WKD8 A0A0Q9X7H2 A0A0Q9WXB7 A0A0Q9X5S6 A0A1I8NGQ2 U5ENN4 A0A1B6KHC7 A0A1S3JFV7 A0A1J1J4A6 A0A1J1J8K3 A0A2I4DAX8 A0A146V7X9 A0A1I8NGQ8 A0A1I8P1T1 A0A1I8NGR3 A0A1I8P1L9
A0A1B6C0X7 A0A1B6DHA6 A0A224XLD3 A0A0K8SEW2 A0A1B6DER4 A0A232F5Q3 A0A1B6G1C8 A0A0N1IT67 A0A0A9WSH7 A0A023F4C5 A0A067RCY7 A0A0A9WSE7 A0A0K8SEV3 A0A0A9WH75 A0A1S4ENM9 A0A2A3EEW1 K7J2A6 K7J2A7 E2ASJ0 A0A2S2Q3F2 E2BZU4 A0A2H8TJY2 A0A0A9WK55 A0A026X1H1 A0A2M4AVN7 A0A0L7RF97 B3P8S1 F5HMI9 B4G619 J9JVX3 B4QZ94 A0A139WKH6 D6WEC4 A0A0Q9VZW4 A0A1A9YUC0 B4JGQ4 A0A0K8U4D6 A0A0K8USR1 A0A0Q9WC59 A0A0Q9VZV6 B4HM48 W8BF57 A0A1A9W5Y9 A0A2C9GQA3 A0A084VVT9 A0A2M4CSE2 A0NGZ6 B3LVH5 A0A0Q9XA77 A0A1W4W1R1 A0A0Q9X046 Q24210 A0A1W4VQ70 Q29AR4 A0A0P8XYE7 A0A3B0JPX6 A0A0P9C8I4 E1JIS7 B4MC57 A0A0R1E448 A0A1W4W370 A0A1W4VQG9 A0A1I8NGR6 A0A0B4KH39 A0A034VQY7 A0A0K8WM54 A0A1I8NGR2 A0A0K8WLW8 A0A0A1X353 A0A0K8VA90 A0A1Y1M581 A0A0K8WKD8 A0A0Q9X7H2 A0A0Q9WXB7 A0A0Q9X5S6 A0A1I8NGQ2 U5ENN4 A0A1B6KHC7 A0A1S3JFV7 A0A1J1J4A6 A0A1J1J8K3 A0A2I4DAX8 A0A146V7X9 A0A1I8NGQ8 A0A1I8P1T1 A0A1I8NGR3 A0A1I8P1L9
EC Number
2.7.11.17
Pubmed
26354079
22118469
19121390
28648823
25401762
25474469
+ More
24845553 20075255 20798317 24508170 17994087 12364791 18362917 19820115 24495485 24438588 9178262 8617233 10731132 12537572 12537569 14687552 15901771 16880127 15632085 12537568 12537573 12537574 16110336 17569856 17569867 17550304 25315136 25348373 25830018 28004739
24845553 20075255 20798317 24508170 17994087 12364791 18362917 19820115 24495485 24438588 9178262 8617233 10731132 12537572 12537569 14687552 15901771 16880127 15632085 12537568 12537573 12537574 16110336 17569856 17569867 17550304 25315136 25348373 25830018 28004739
EMBL
KQ460324
KPJ15842.1
AGBW02011024
OWR47350.1
BABH01025906
KQ459249
+ More
KPJ02406.1 NWSH01001543 PCG70905.1 PCG70906.1 GEDC01030126 JAS07172.1 GEDC01012231 JAS25067.1 GFTR01007585 JAW08841.1 GBRD01014003 JAG51823.1 GEDC01013168 JAS24130.1 NNAY01000914 OXU25942.1 GECZ01013521 JAS56248.1 KQ435859 KOX70502.1 GBHO01035804 JAG07800.1 GBBI01002351 JAC16361.1 KK852547 KDR21602.1 GBHO01032900 JAG10704.1 GBRD01014004 JAG51822.1 GBHO01035802 JAG07802.1 KZ288272 PBC29739.1 AAZX01001591 AAZX01006227 AAZX01008245 GL442313 EFN63587.1 GGMS01002948 MBY72151.1 GL451689 EFN78780.1 GFXV01002177 MBW13982.1 GBHO01035803 JAG07801.1 KK107037 EZA61943.1 GGFK01011532 MBW44853.1 KQ414607 KOC69489.1 CH954182 EDV54235.1 AAAB01008987 EGK97511.1 CH479179 EDW23778.1 ABLF02026617 CM000364 EDX13830.1 KQ971326 KYB28489.1 EEZ99889.2 CH940656 KRF78394.1 CCAG010002878 CH916369 EDV92658.1 GDHF01030891 JAI21423.1 GDHF01022756 JAI29558.1 KRF78395.1 KRF78393.1 CH480815 EDW43096.1 GAMC01009233 JAB97322.1 APCN01000909 ATLV01017289 ATLV01017290 ATLV01017291 ATLV01017292 ATLV01017293 ATLV01017294 ATLV01017295 KE525161 KFB42083.1 GGFL01004072 MBW68250.1 EAU75645.3 CH902617 EDV42545.2 CH933806 KRG01502.1 KRG01500.1 U53190 X94264 AE014297 AY094916 BT003532 BT003550 BT046172 CM000070 EAL27285.3 KPU79785.1 OUUW01000007 SPP82953.1 KPU79787.1 ACZ94977.1 EDW58678.2 CM000160 KRK04027.1 AGB96191.1 GAKP01014063 JAC44889.1 GDHF01000170 JAI52144.1 GDHF01000156 JAI52158.1 GBXI01010360 GBXI01008548 JAD03932.1 JAD05744.1 GDHF01016503 JAI35811.1 GEZM01042960 JAV79445.1 GDHF01000790 JAI51524.1 CH964272 KRG00088.1 KRG00089.1 KRG00087.1 GANO01000524 JAB59347.1 GEBQ01029121 JAT10856.1 CVRI01000070 CRL07232.1 CRL07233.1 GCES01072810 JAR13513.1
KPJ02406.1 NWSH01001543 PCG70905.1 PCG70906.1 GEDC01030126 JAS07172.1 GEDC01012231 JAS25067.1 GFTR01007585 JAW08841.1 GBRD01014003 JAG51823.1 GEDC01013168 JAS24130.1 NNAY01000914 OXU25942.1 GECZ01013521 JAS56248.1 KQ435859 KOX70502.1 GBHO01035804 JAG07800.1 GBBI01002351 JAC16361.1 KK852547 KDR21602.1 GBHO01032900 JAG10704.1 GBRD01014004 JAG51822.1 GBHO01035802 JAG07802.1 KZ288272 PBC29739.1 AAZX01001591 AAZX01006227 AAZX01008245 GL442313 EFN63587.1 GGMS01002948 MBY72151.1 GL451689 EFN78780.1 GFXV01002177 MBW13982.1 GBHO01035803 JAG07801.1 KK107037 EZA61943.1 GGFK01011532 MBW44853.1 KQ414607 KOC69489.1 CH954182 EDV54235.1 AAAB01008987 EGK97511.1 CH479179 EDW23778.1 ABLF02026617 CM000364 EDX13830.1 KQ971326 KYB28489.1 EEZ99889.2 CH940656 KRF78394.1 CCAG010002878 CH916369 EDV92658.1 GDHF01030891 JAI21423.1 GDHF01022756 JAI29558.1 KRF78395.1 KRF78393.1 CH480815 EDW43096.1 GAMC01009233 JAB97322.1 APCN01000909 ATLV01017289 ATLV01017290 ATLV01017291 ATLV01017292 ATLV01017293 ATLV01017294 ATLV01017295 KE525161 KFB42083.1 GGFL01004072 MBW68250.1 EAU75645.3 CH902617 EDV42545.2 CH933806 KRG01502.1 KRG01500.1 U53190 X94264 AE014297 AY094916 BT003532 BT003550 BT046172 CM000070 EAL27285.3 KPU79785.1 OUUW01000007 SPP82953.1 KPU79787.1 ACZ94977.1 EDW58678.2 CM000160 KRK04027.1 AGB96191.1 GAKP01014063 JAC44889.1 GDHF01000170 JAI52144.1 GDHF01000156 JAI52158.1 GBXI01010360 GBXI01008548 JAD03932.1 JAD05744.1 GDHF01016503 JAI35811.1 GEZM01042960 JAV79445.1 GDHF01000790 JAI51524.1 CH964272 KRG00088.1 KRG00089.1 KRG00087.1 GANO01000524 JAB59347.1 GEBQ01029121 JAT10856.1 CVRI01000070 CRL07232.1 CRL07233.1 GCES01072810 JAR13513.1
Proteomes
UP000053240
UP000007151
UP000005204
UP000053268
UP000218220
UP000215335
+ More
UP000053105 UP000027135 UP000079169 UP000242457 UP000002358 UP000000311 UP000008237 UP000053097 UP000053825 UP000008711 UP000007062 UP000008744 UP000007819 UP000000304 UP000007266 UP000008792 UP000092444 UP000001070 UP000001292 UP000091820 UP000075840 UP000030765 UP000007801 UP000009192 UP000192221 UP000000803 UP000001819 UP000268350 UP000002282 UP000095301 UP000007798 UP000085678 UP000183832 UP000192220 UP000095300
UP000053105 UP000027135 UP000079169 UP000242457 UP000002358 UP000000311 UP000008237 UP000053097 UP000053825 UP000008711 UP000007062 UP000008744 UP000007819 UP000000304 UP000007266 UP000008792 UP000092444 UP000001070 UP000001292 UP000091820 UP000075840 UP000030765 UP000007801 UP000009192 UP000192221 UP000000803 UP000001819 UP000268350 UP000002282 UP000095301 UP000007798 UP000085678 UP000183832 UP000192220 UP000095300
Pfam
Interpro
IPR011009
Kinase-like_dom_sf
+ More
IPR036892 L27_dom_sf
IPR014775 L27_C
IPR000719 Prot_kinase_dom
IPR004172 L27_dom
IPR008145 GK/Ca_channel_bsu
IPR001452 SH3_domain
IPR008144 Guanylate_kin-like_dom
IPR036028 SH3-like_dom_sf
IPR027417 P-loop_NTPase
IPR001478 PDZ
IPR036034 PDZ_sf
IPR020590 Guanylate_kinase_CS
IPR035473 CASK_SH3
IPR036892 L27_dom_sf
IPR014775 L27_C
IPR000719 Prot_kinase_dom
IPR004172 L27_dom
IPR008145 GK/Ca_channel_bsu
IPR001452 SH3_domain
IPR008144 Guanylate_kin-like_dom
IPR036028 SH3-like_dom_sf
IPR027417 P-loop_NTPase
IPR001478 PDZ
IPR036034 PDZ_sf
IPR020590 Guanylate_kinase_CS
IPR035473 CASK_SH3
SUPFAM
CDD
ProteinModelPortal
A0A0N0PD55
A0A212F0U0
H9J9V3
A0A194QG32
A0A2A4JFP8
A0A2A4JGK0
+ More
A0A1B6C0X7 A0A1B6DHA6 A0A224XLD3 A0A0K8SEW2 A0A1B6DER4 A0A232F5Q3 A0A1B6G1C8 A0A0N1IT67 A0A0A9WSH7 A0A023F4C5 A0A067RCY7 A0A0A9WSE7 A0A0K8SEV3 A0A0A9WH75 A0A1S4ENM9 A0A2A3EEW1 K7J2A6 K7J2A7 E2ASJ0 A0A2S2Q3F2 E2BZU4 A0A2H8TJY2 A0A0A9WK55 A0A026X1H1 A0A2M4AVN7 A0A0L7RF97 B3P8S1 F5HMI9 B4G619 J9JVX3 B4QZ94 A0A139WKH6 D6WEC4 A0A0Q9VZW4 A0A1A9YUC0 B4JGQ4 A0A0K8U4D6 A0A0K8USR1 A0A0Q9WC59 A0A0Q9VZV6 B4HM48 W8BF57 A0A1A9W5Y9 A0A2C9GQA3 A0A084VVT9 A0A2M4CSE2 A0NGZ6 B3LVH5 A0A0Q9XA77 A0A1W4W1R1 A0A0Q9X046 Q24210 A0A1W4VQ70 Q29AR4 A0A0P8XYE7 A0A3B0JPX6 A0A0P9C8I4 E1JIS7 B4MC57 A0A0R1E448 A0A1W4W370 A0A1W4VQG9 A0A1I8NGR6 A0A0B4KH39 A0A034VQY7 A0A0K8WM54 A0A1I8NGR2 A0A0K8WLW8 A0A0A1X353 A0A0K8VA90 A0A1Y1M581 A0A0K8WKD8 A0A0Q9X7H2 A0A0Q9WXB7 A0A0Q9X5S6 A0A1I8NGQ2 U5ENN4 A0A1B6KHC7 A0A1S3JFV7 A0A1J1J4A6 A0A1J1J8K3 A0A2I4DAX8 A0A146V7X9 A0A1I8NGQ8 A0A1I8P1T1 A0A1I8NGR3 A0A1I8P1L9
A0A1B6C0X7 A0A1B6DHA6 A0A224XLD3 A0A0K8SEW2 A0A1B6DER4 A0A232F5Q3 A0A1B6G1C8 A0A0N1IT67 A0A0A9WSH7 A0A023F4C5 A0A067RCY7 A0A0A9WSE7 A0A0K8SEV3 A0A0A9WH75 A0A1S4ENM9 A0A2A3EEW1 K7J2A6 K7J2A7 E2ASJ0 A0A2S2Q3F2 E2BZU4 A0A2H8TJY2 A0A0A9WK55 A0A026X1H1 A0A2M4AVN7 A0A0L7RF97 B3P8S1 F5HMI9 B4G619 J9JVX3 B4QZ94 A0A139WKH6 D6WEC4 A0A0Q9VZW4 A0A1A9YUC0 B4JGQ4 A0A0K8U4D6 A0A0K8USR1 A0A0Q9WC59 A0A0Q9VZV6 B4HM48 W8BF57 A0A1A9W5Y9 A0A2C9GQA3 A0A084VVT9 A0A2M4CSE2 A0NGZ6 B3LVH5 A0A0Q9XA77 A0A1W4W1R1 A0A0Q9X046 Q24210 A0A1W4VQ70 Q29AR4 A0A0P8XYE7 A0A3B0JPX6 A0A0P9C8I4 E1JIS7 B4MC57 A0A0R1E448 A0A1W4W370 A0A1W4VQG9 A0A1I8NGR6 A0A0B4KH39 A0A034VQY7 A0A0K8WM54 A0A1I8NGR2 A0A0K8WLW8 A0A0A1X353 A0A0K8VA90 A0A1Y1M581 A0A0K8WKD8 A0A0Q9X7H2 A0A0Q9WXB7 A0A0Q9X5S6 A0A1I8NGQ2 U5ENN4 A0A1B6KHC7 A0A1S3JFV7 A0A1J1J4A6 A0A1J1J8K3 A0A2I4DAX8 A0A146V7X9 A0A1I8NGQ8 A0A1I8P1T1 A0A1I8NGR3 A0A1I8P1L9
PDB
3TAC
E-value=2.65704e-125,
Score=1149
Ontologies
GO
GO:0005524
GO:0004683
GO:0004672
GO:0004674
GO:0007615
GO:0008049
GO:0042043
GO:0008582
GO:0031594
GO:0007616
GO:0040011
GO:0005886
GO:2000331
GO:0040012
GO:0005516
GO:0072375
GO:0007269
GO:0061174
GO:0007163
GO:0048149
GO:0007274
GO:0048488
GO:0008344
GO:0007298
GO:0016080
GO:0007628
GO:0046928
GO:0016081
GO:1900244
GO:1900073
GO:0005515
GO:0006468
GO:0016020
GO:0051103
GO:0008270
GO:0042555
GO:0048384
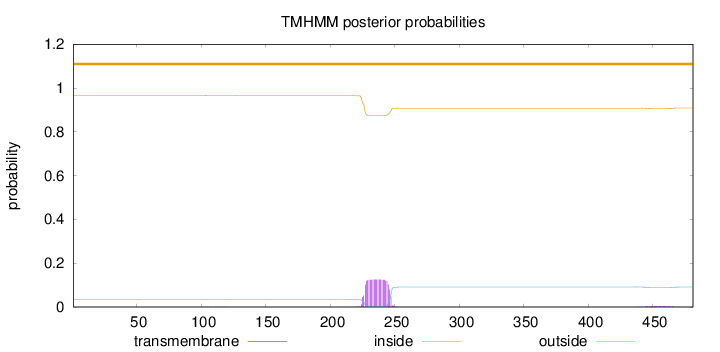
Topology
Subcellular location
Cell membrane
Length:
481
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.65539
Exp number, first 60 AAs:
0.000990000000000001
Total prob of N-in:
0.03474
outside
1 - 481
Population Genetic Test Statistics
Pi
220.230935
Theta
155.853641
Tajima's D
1.028479
CLR
0.294078
CSRT
0.665416729163542
Interpretation
Uncertain
