Gene
KWMTBOMO03559
Pre Gene Modal
BGIBMGA006297
Annotation
PREDICTED:_kal-1_protein_isoform_X1_[Bombyx_mori]
Full name
Anosmin-1
Alternative Name
Kallmann syndrome protein homolog
Location in the cell
Nuclear Reliability : 2.073
Sequence
CDS
ATGCTGATGAGACTGTTGACGGTGATGGTGGTGGCGTCTCTCGTTCCTGCGATCCAAGCGAGGGCGCGGAGATACTCGAGAATGCAACCTAATGCTTTATCAAAGACAAGATGCGATCTGATGTGCTTTGACAGGGATAAGGAAACTAAAGGAACGTGTCGTTCCCAATGTCGGAATCAAGAGCACAAGCCGGGAACATGCCCCGTTTCGGACACGCCGAAATGGGAAGCGGCGTGCGTGCAGGCCTGCAACTCCGACTCCCAATGCGACGGCACACAGAGGTGCTGTCACCACGGATGCGGATCTACTTGCAGTGAACCTTTAGATTTGTTGACTTTACCAGGTCTTCCAGCCGTTCCTACAATGGAAGAACCAAAGGAGAAGCGACGAGCCGTCGTTTTGCGCTGGTCAGACGGCGTAGGAGATACAGCGAGGGCTGTTCCAGGACGAGTCCTTTACCTGCTTGAGGAACAACATCATTTGGGCCCTAAATATGAGCAGTCTAGGCTTGGCGATTGGAATCTCATGTTACGTACTAACAGGACAAAGGTTTCGCTGAGGAATCTGTTGAAACCAGGTCGCTGGTATCGGTTCCGTGTTGCTGCTATTAGCGCAGCTGGTACACGAGGGTTCTCCGATCCCAGTCCTCCATTTACACCACGTCGTGGACCCCGTCCTCCTCCTATGCCGAAGAAATTGAGAGTCCGACCAATGAGAATGGATAACGGAACGATGACAGTAAGACTTGAATGGAAGGAGCCTCGTTCTGATCTGCCAGTGATGCGCTACAAGGTATTCTGGAGTAGACGAGTAAGAGGATTGGGCGGAGAGCTAGATTCCGTTCTTGTCAATCATCAGACAGTTTCAAAGGATCAAAACCACATCGAAATAAAAGATTTACAACCGAATTCTATGTACTTTTTGCAAGTTCAAACGATCAGTCAATTTGGTTTGGGTAAATTGCGAAGCGATAAGGCATCTGTATTTTACAACACGACTGGTTCTGTAGGAAATGATTCTGTACCAGAATCTTTAGTCAGACGTGACAAATATATAAAGGGTTTGAAGTTGAATAAAATAGTTTGGAACAATCAAAGACTGAAAGCGAGAATTTCATGGGAATCAGTGCCTAGTGGAAACAGTGGCGGAAGGTCACAAGAGAGATATTACGTCCATTGGAAAACTATAAAATGCAATAAAACAGATAAACCCATGAAAGACTTAGCGGCGACCACAGCGCAATCTACGTTTGAACTCTATGAATTGGATTACCACTGCAGCTATAAAGTAAATGTGAACAGATCCTGGAAGAATAAAATACCAGAATCAGAGCTCGTTATAACAATTCCTGGATGTCAGTATTTTAAAAGAAAAGTTAATGTTACAGCCATAACGTGTGACTCGTAA
Protein
MLMRLLTVMVVASLVPAIQARARRYSRMQPNALSKTRCDLMCFDRDKETKGTCRSQCRNQEHKPGTCPVSDTPKWEAACVQACNSDSQCDGTQRCCHHGCGSTCSEPLDLLTLPGLPAVPTMEEPKEKRRAVVLRWSDGVGDTARAVPGRVLYLLEEQHHLGPKYEQSRLGDWNLMLRTNRTKVSLRNLLKPGRWYRFRVAAISAAGTRGFSDPSPPFTPRRGPRPPPMPKKLRVRPMRMDNGTMTVRLEWKEPRSDLPVMRYKVFWSRRVRGLGGELDSVLVNHQTVSKDQNHIEIKDLQPNSMYFLQVQTISQFGLGKLRSDKASVFYNTTGSVGNDSVPESLVRRDKYIKGLKLNKIVWNNQRLKARISWESVPSGNSGGRSQERYYVHWKTIKCNKTDKPMKDLAATTAQSTFELYELDYHCSYKVNVNRSWKNKIPESELVITIPGCQYFKRKVNVTAITCDS
Summary
Keywords
Cell adhesion
Disulfide bond
Glycoprotein
Protease inhibitor
Repeat
Serine protease inhibitor
Signal
Feature
chain Anosmin-1
Uniprot
Q8WS93
A0A2A4J0B0
A0A0N1IFD7
A0A194QAA3
A0A212F0X1
A0A0L7LMQ2
+ More
A0A2H1VPX8 A0A139WM55 A0A3L8DU14 A0A026X0E2 A0A088A9B5 A0A2S2QI92 A0A158NUI3 A0A1B6DTF5 F4X175 A0A195AY95 A0A151XHF4 E2AWQ7 A0A154PM94 A0A195DUP7 A0A0C9PK66 A0A0C9QU84 J9JR45 E2BLE3 A0A2H8TYV4 A0A2S2NWT7 A0A195ESE5 H3A9M1 C3ZEN5 B0W121 A0A1Q3FHS2 D2CFV1 A0A1Q3FHP4 F7FF48 G1K8I7 F6S024 A0A1S4F2B2 Q17HU5 Q17HU4 A0A093I3M1 A0A1L8HDG5 A0A091PHS3 A0A1S3H5D2 A0A1L8H4Q6 A9JRI8 A0A091TLP0 A0A182GLS8 F7AUG0 A0A182R834 F6V374 G5E7N3 A0A093Q3Y6 A0A151N0M3 A0A093BWH1 A0A182JNJ8 A0A383ZGT6 A0A2M4ASJ7 A0A0A0AIV7 A0A091MHF9 A0A091FTG6 V9KKG4 A0A087VCI4 A0A087RII3 A0A091NL59 A0A2I0ULT4 A0A091SM39 U3IA36 A0A1V4JCR5 A0A091V7F5 A0A091PCI8 A0A182NKX1 A0A182LRG8 A0A0Q3M8V8 A0A091I2B1 A0A093PBE2 R7UKM7 A0A093JAV1 G1NPF0 Q90369 A0A3P8VL27 A0A091J7B2 H0ZDZ2 R0K340 A0A091FHC7 U3JPP4 A0A182J219
A0A2H1VPX8 A0A139WM55 A0A3L8DU14 A0A026X0E2 A0A088A9B5 A0A2S2QI92 A0A158NUI3 A0A1B6DTF5 F4X175 A0A195AY95 A0A151XHF4 E2AWQ7 A0A154PM94 A0A195DUP7 A0A0C9PK66 A0A0C9QU84 J9JR45 E2BLE3 A0A2H8TYV4 A0A2S2NWT7 A0A195ESE5 H3A9M1 C3ZEN5 B0W121 A0A1Q3FHS2 D2CFV1 A0A1Q3FHP4 F7FF48 G1K8I7 F6S024 A0A1S4F2B2 Q17HU5 Q17HU4 A0A093I3M1 A0A1L8HDG5 A0A091PHS3 A0A1S3H5D2 A0A1L8H4Q6 A9JRI8 A0A091TLP0 A0A182GLS8 F7AUG0 A0A182R834 F6V374 G5E7N3 A0A093Q3Y6 A0A151N0M3 A0A093BWH1 A0A182JNJ8 A0A383ZGT6 A0A2M4ASJ7 A0A0A0AIV7 A0A091MHF9 A0A091FTG6 V9KKG4 A0A087VCI4 A0A087RII3 A0A091NL59 A0A2I0ULT4 A0A091SM39 U3IA36 A0A1V4JCR5 A0A091V7F5 A0A091PCI8 A0A182NKX1 A0A182LRG8 A0A0Q3M8V8 A0A091I2B1 A0A093PBE2 R7UKM7 A0A093JAV1 G1NPF0 Q90369 A0A3P8VL27 A0A091J7B2 H0ZDZ2 R0K340 A0A091FHC7 U3JPP4 A0A182J219
Pubmed
EMBL
AF342987
AAL73339.1
NWSH01004146
PCG65431.1
KQ460324
KPJ15843.1
+ More
KQ459249 KPJ02407.1 AGBW02011024 OWR47351.1 JTDY01000556 KOB76690.1 ODYU01003744 SOQ42901.1 KQ971312 KYB29099.1 QOIP01000004 RLU23861.1 KK107046 EZA61765.1 GGMS01007699 MBY76902.1 ADTU01026478 ADTU01026479 ADTU01026480 ADTU01026481 ADTU01026482 ADTU01026483 GEDC01008341 JAS28957.1 GL888527 EGI59811.1 KQ976711 KYM77012.1 KQ982138 KYQ59640.1 GL443403 EFN62133.1 KQ434946 KZC12340.1 KQ980390 KYN16279.1 GBYB01001413 JAG71180.1 GBYB01007279 JAG77046.1 ABLF02040114 GL449017 EFN83495.1 GFXV01007641 MBW19446.1 GGMR01009016 MBY21635.1 KQ981989 KYN31148.1 AFYH01209423 AFYH01209424 AFYH01209425 AFYH01209426 AFYH01209427 AFYH01209428 AFYH01209429 AFYH01209430 GG666612 EEN49191.1 DS231819 EDS42838.1 GFDL01007929 JAV27116.1 DQ991133 ABL74507.1 GFDL01007968 JAV27077.1 CH477245 EAT46247.1 EAT46248.1 KL206793 KFV85802.1 CM004468 OCT94071.1 KK673305 KFQ06798.1 CM004469 OCT91079.1 BC155677 BC170858 AAI55678.1 AAI70858.1 KK458425 KFQ77738.1 JXUM01073053 JXUM01073054 KQ562754 KXJ75197.1 AAMC01087004 AAMC01087005 AAMC01087006 AAMC01087007 AAMC01087008 AAMC01087009 AAMC01087010 AAMC01087011 AAMC01087012 KL671660 KFW83361.1 AKHW03004278 KYO30302.1 KL450501 KFV05194.1 GGFK01010455 MBW43776.1 KL871948 KGL94081.1 KK825724 KFP72605.1 KL447348 KFO72484.1 JW866102 AFO98619.1 KL488263 KFO10326.1 KL226387 KFM13287.1 KL385684 KFP89822.1 KZ505692 PKU46999.1 KK478335 KFQ59709.1 ADON01047117 ADON01047118 ADON01047119 ADON01047120 ADON01047121 ADON01047122 LSYS01008075 OPJ69557.1 KL410696 KFQ98302.1 KK656681 KFQ05355.1 AXCM01000812 LMAW01002627 KQK78864.1 KL218032 KFP01565.1 KL225535 KFW73736.1 AMQN01008197 KB302404 ELU04353.1 KK573574 KFW09012.1 L13976 KK501586 KFP16487.1 ABQF01012569 ABQF01012570 ABQF01012571 KB742810 EOB04062.1 KK718990 KFO60850.1 AGTO01000472
KQ459249 KPJ02407.1 AGBW02011024 OWR47351.1 JTDY01000556 KOB76690.1 ODYU01003744 SOQ42901.1 KQ971312 KYB29099.1 QOIP01000004 RLU23861.1 KK107046 EZA61765.1 GGMS01007699 MBY76902.1 ADTU01026478 ADTU01026479 ADTU01026480 ADTU01026481 ADTU01026482 ADTU01026483 GEDC01008341 JAS28957.1 GL888527 EGI59811.1 KQ976711 KYM77012.1 KQ982138 KYQ59640.1 GL443403 EFN62133.1 KQ434946 KZC12340.1 KQ980390 KYN16279.1 GBYB01001413 JAG71180.1 GBYB01007279 JAG77046.1 ABLF02040114 GL449017 EFN83495.1 GFXV01007641 MBW19446.1 GGMR01009016 MBY21635.1 KQ981989 KYN31148.1 AFYH01209423 AFYH01209424 AFYH01209425 AFYH01209426 AFYH01209427 AFYH01209428 AFYH01209429 AFYH01209430 GG666612 EEN49191.1 DS231819 EDS42838.1 GFDL01007929 JAV27116.1 DQ991133 ABL74507.1 GFDL01007968 JAV27077.1 CH477245 EAT46247.1 EAT46248.1 KL206793 KFV85802.1 CM004468 OCT94071.1 KK673305 KFQ06798.1 CM004469 OCT91079.1 BC155677 BC170858 AAI55678.1 AAI70858.1 KK458425 KFQ77738.1 JXUM01073053 JXUM01073054 KQ562754 KXJ75197.1 AAMC01087004 AAMC01087005 AAMC01087006 AAMC01087007 AAMC01087008 AAMC01087009 AAMC01087010 AAMC01087011 AAMC01087012 KL671660 KFW83361.1 AKHW03004278 KYO30302.1 KL450501 KFV05194.1 GGFK01010455 MBW43776.1 KL871948 KGL94081.1 KK825724 KFP72605.1 KL447348 KFO72484.1 JW866102 AFO98619.1 KL488263 KFO10326.1 KL226387 KFM13287.1 KL385684 KFP89822.1 KZ505692 PKU46999.1 KK478335 KFQ59709.1 ADON01047117 ADON01047118 ADON01047119 ADON01047120 ADON01047121 ADON01047122 LSYS01008075 OPJ69557.1 KL410696 KFQ98302.1 KK656681 KFQ05355.1 AXCM01000812 LMAW01002627 KQK78864.1 KL218032 KFP01565.1 KL225535 KFW73736.1 AMQN01008197 KB302404 ELU04353.1 KK573574 KFW09012.1 L13976 KK501586 KFP16487.1 ABQF01012569 ABQF01012570 ABQF01012571 KB742810 EOB04062.1 KK718990 KFO60850.1 AGTO01000472
Proteomes
UP000218220
UP000053240
UP000053268
UP000007151
UP000037510
UP000007266
+ More
UP000279307 UP000053097 UP000005203 UP000005205 UP000007755 UP000078540 UP000075809 UP000000311 UP000076502 UP000078492 UP000007819 UP000008237 UP000078541 UP000008672 UP000001554 UP000002320 UP000002280 UP000001646 UP000002279 UP000008820 UP000053584 UP000186698 UP000085678 UP000069940 UP000249989 UP000008143 UP000075900 UP000001645 UP000053258 UP000050525 UP000075881 UP000261681 UP000053858 UP000053760 UP000053286 UP000016666 UP000190648 UP000053283 UP000075884 UP000075883 UP000051836 UP000054308 UP000054081 UP000014760 UP000265120 UP000053119 UP000007754 UP000052976 UP000016665 UP000075880
UP000279307 UP000053097 UP000005203 UP000005205 UP000007755 UP000078540 UP000075809 UP000000311 UP000076502 UP000078492 UP000007819 UP000008237 UP000078541 UP000008672 UP000001554 UP000002320 UP000002280 UP000001646 UP000002279 UP000008820 UP000053584 UP000186698 UP000085678 UP000069940 UP000249989 UP000008143 UP000075900 UP000001645 UP000053258 UP000050525 UP000075881 UP000261681 UP000053858 UP000053760 UP000053286 UP000016666 UP000190648 UP000053283 UP000075884 UP000075883 UP000051836 UP000054308 UP000054081 UP000014760 UP000265120 UP000053119 UP000007754 UP000052976 UP000016665 UP000075880
PRIDE
Pfam
Interpro
IPR013783
Ig-like_fold
+ More
IPR008197 WAP_dom
IPR003961 FN3_dom
IPR036116 FN3_sf
IPR036645 Elafin-like_sf
IPR034016 M1_APN-typ
IPR001930 Peptidase_M1
IPR033581 APN2
IPR042097 Aminopeptidase_N-like_N
IPR014782 Peptidase_M1_dom
IPR024571 ERAP1-like_C_dom
IPR040957 Anosmin-1_Cys_box
IPR036179 Ig-like_dom_sf
IPR003598 Ig_sub2
IPR013098 Ig_I-set
IPR007110 Ig-like_dom
IPR003599 Ig_sub
IPR008197 WAP_dom
IPR003961 FN3_dom
IPR036116 FN3_sf
IPR036645 Elafin-like_sf
IPR034016 M1_APN-typ
IPR001930 Peptidase_M1
IPR033581 APN2
IPR042097 Aminopeptidase_N-like_N
IPR014782 Peptidase_M1_dom
IPR024571 ERAP1-like_C_dom
IPR040957 Anosmin-1_Cys_box
IPR036179 Ig-like_dom_sf
IPR003598 Ig_sub2
IPR013098 Ig_I-set
IPR007110 Ig-like_dom
IPR003599 Ig_sub
Gene 3D
ProteinModelPortal
Q8WS93
A0A2A4J0B0
A0A0N1IFD7
A0A194QAA3
A0A212F0X1
A0A0L7LMQ2
+ More
A0A2H1VPX8 A0A139WM55 A0A3L8DU14 A0A026X0E2 A0A088A9B5 A0A2S2QI92 A0A158NUI3 A0A1B6DTF5 F4X175 A0A195AY95 A0A151XHF4 E2AWQ7 A0A154PM94 A0A195DUP7 A0A0C9PK66 A0A0C9QU84 J9JR45 E2BLE3 A0A2H8TYV4 A0A2S2NWT7 A0A195ESE5 H3A9M1 C3ZEN5 B0W121 A0A1Q3FHS2 D2CFV1 A0A1Q3FHP4 F7FF48 G1K8I7 F6S024 A0A1S4F2B2 Q17HU5 Q17HU4 A0A093I3M1 A0A1L8HDG5 A0A091PHS3 A0A1S3H5D2 A0A1L8H4Q6 A9JRI8 A0A091TLP0 A0A182GLS8 F7AUG0 A0A182R834 F6V374 G5E7N3 A0A093Q3Y6 A0A151N0M3 A0A093BWH1 A0A182JNJ8 A0A383ZGT6 A0A2M4ASJ7 A0A0A0AIV7 A0A091MHF9 A0A091FTG6 V9KKG4 A0A087VCI4 A0A087RII3 A0A091NL59 A0A2I0ULT4 A0A091SM39 U3IA36 A0A1V4JCR5 A0A091V7F5 A0A091PCI8 A0A182NKX1 A0A182LRG8 A0A0Q3M8V8 A0A091I2B1 A0A093PBE2 R7UKM7 A0A093JAV1 G1NPF0 Q90369 A0A3P8VL27 A0A091J7B2 H0ZDZ2 R0K340 A0A091FHC7 U3JPP4 A0A182J219
A0A2H1VPX8 A0A139WM55 A0A3L8DU14 A0A026X0E2 A0A088A9B5 A0A2S2QI92 A0A158NUI3 A0A1B6DTF5 F4X175 A0A195AY95 A0A151XHF4 E2AWQ7 A0A154PM94 A0A195DUP7 A0A0C9PK66 A0A0C9QU84 J9JR45 E2BLE3 A0A2H8TYV4 A0A2S2NWT7 A0A195ESE5 H3A9M1 C3ZEN5 B0W121 A0A1Q3FHS2 D2CFV1 A0A1Q3FHP4 F7FF48 G1K8I7 F6S024 A0A1S4F2B2 Q17HU5 Q17HU4 A0A093I3M1 A0A1L8HDG5 A0A091PHS3 A0A1S3H5D2 A0A1L8H4Q6 A9JRI8 A0A091TLP0 A0A182GLS8 F7AUG0 A0A182R834 F6V374 G5E7N3 A0A093Q3Y6 A0A151N0M3 A0A093BWH1 A0A182JNJ8 A0A383ZGT6 A0A2M4ASJ7 A0A0A0AIV7 A0A091MHF9 A0A091FTG6 V9KKG4 A0A087VCI4 A0A087RII3 A0A091NL59 A0A2I0ULT4 A0A091SM39 U3IA36 A0A1V4JCR5 A0A091V7F5 A0A091PCI8 A0A182NKX1 A0A182LRG8 A0A0Q3M8V8 A0A091I2B1 A0A093PBE2 R7UKM7 A0A093JAV1 G1NPF0 Q90369 A0A3P8VL27 A0A091J7B2 H0ZDZ2 R0K340 A0A091FHC7 U3JPP4 A0A182J219
PDB
1ZLG
E-value=1.01756e-31,
Score=342
Ontologies
GO
PANTHER
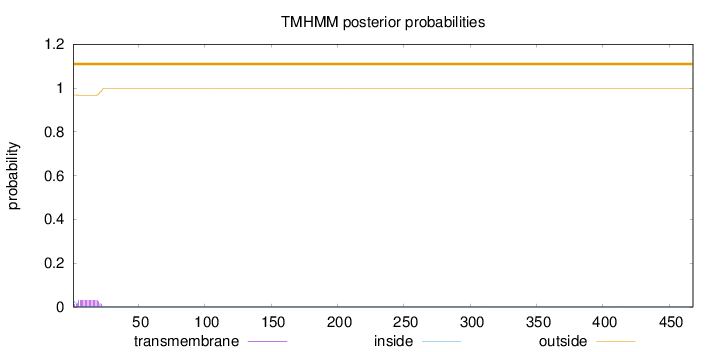
Topology
Subcellular location
Cell surface
SignalP
Position: 1 - 20,
Likelihood: 0.992338
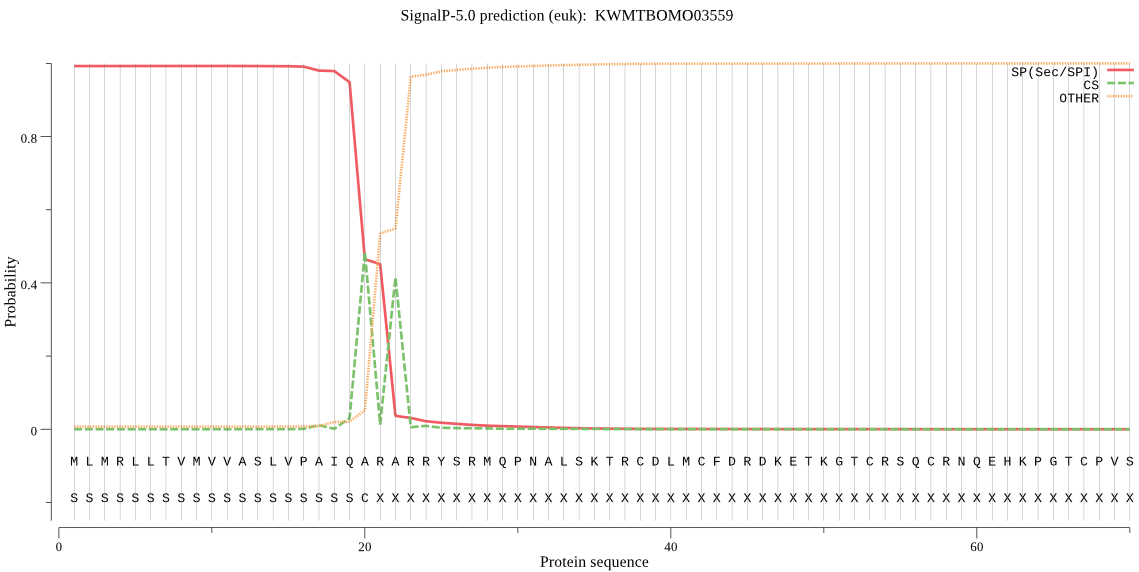
Length:
468
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.57269
Exp number, first 60 AAs:
0.57178
Total prob of N-in:
0.03226
outside
1 - 468
Population Genetic Test Statistics
Pi
242.050248
Theta
175.206608
Tajima's D
1.35384
CLR
0.028967
CSRT
0.753312334383281
Interpretation
Uncertain
