Gene
KWMTBOMO03557
Pre Gene Modal
BGIBMGA006296
Annotation
PREDICTED:_lopap_[Papilio_machaon]
Location in the cell
Extracellular Reliability : 1.146 PlasmaMembrane Reliability : 1.787
Sequence
CDS
ATGGTGAAGTTTTTAATGTTTATTGTGTTTTTGTTATTTTCGAACGTGGTGATCGGAAAAATTGTAAACAAAAGAAAGTGCCCAGAGGTGCGAGCTGTACCACACTTCGATTTGCCCGGGATACTGGGAGACTGGTATGTGGTTGAGTACTACGCGAGTGCTGAAGAAGCGCTCTCATACAGTTGTATGAAAGCCGTGTTTACTCAGGACGATCACGTACCCGCCATAGATGGTACAATAGAGTGGACGCCAGGAGTGACCATGAACTTTACGTACCGGTTCGCGGACGACCCCATAGGCGAGACACTCTTCGGAAACATTACCTGGAAGATCGATCTGAACCAGCCTGCGCACTGGACACACGCTGAAAGCACATATGACGGCATTTACAATACGTACGTGATAGACACAGAATATAAAACTTGGGCGTTACTGCTGCACTGCGCGGAGCAGACGGAAGGCGCTCGTTACCTCACCGCATTCGTTCTCTCCAGGACAACGAAACTGCAGAAGAACGTGATGGCGTACCTGCGAGACAAGCTGCCAAGATTCGAAGTGGACATCAACTATGTGTTCGCGATCCCTCACGACGACTGCGCCGTGAAACCCCCAAACCCTAACTTCGGACCCTTGATGCTGGATGCCCAGAGCAAGAAACTGATAAGACCATCTGTCAGTTACAAAGTCTTCGGTCATTAA
Protein
MVKFLMFIVFLLFSNVVIGKIVNKRKCPEVRAVPHFDLPGILGDWYVVEYYASAEEALSYSCMKAVFTQDDHVPAIDGTIEWTPGVTMNFTYRFADDPIGETLFGNITWKIDLNQPAHWTHAESTYDGIYNTYVIDTEYKTWALLLHCAEQTEGARYLTAFVLSRTTKLQKNVMAYLRDKLPRFEVDINYVFAIPHDDCAVKPPNPNFGPLMLDAQSKKLIRPSVSYKVFGH
Summary
Similarity
Belongs to the calycin superfamily. Lipocalin family.
Uniprot
H9J9V1
A0A2H1WKQ0
A0A0N1I8Q8
A0A212F0V5
A0A2A4J0M7
A0A194QAF1
+ More
A0A067QUM8 A0A1W4WMU8 N6T527 A0A088AQU4 A0A2A3EDB8 A0A1B6HLV1 A0A1B6F8X6 D6WEM3 A0A1Q3FS32 K7J2A5 A0A232F565 E2BVR3 A0A026X0W6 A0A195EZA2 A0A182GCV2 A0A1Y1MUQ1 A0A151WZF6 F4X2A3 E9IHT8 A0A151J6S3 A0A158P389 A0A154PE27 A0A023EKX8 A0A195CEV6 Q17HX8 A0A1I8NX44 B0XGR3 A0A1B6LP33 E0VTZ2 A0A1A9WCJ5 W8BSM6 A0A1I8MGX2 A0A1S4F1Y0 A0A182PM69 A0A0K8W233 A0A034W1U3 A0A0C9R2X2 A0A2M4DP35 A0A1A9XDV9 A0A0L0BRE4 A0A0M4EUT1 B4L1R2 A0A182V7F6 A0A0P4VLH9 T1HNH3 A0A182N0Y6 B4MEJ2 A0A084VIU5 A0A182XY43 A0A0V0GCT9 A0A1S3DGM5 A0A182RJ22 A0A182M8Y1 A0A182FM96 A0A182VVB8 A0A182J456 B4JM62 A0A3B0K5N9 A0A195BVF1 A0A182TQ91 A0A182X5B8 A0A0N0BDQ0 A0A1B0CVH5 A0A2S2RAM8 B4H3R8 W5JPG1 Q29JB5 A0A0A9WEG4 A0A182I9L8 A0A224XWG2 A0A182SR27 B3NU89 Q9VYU5 Q962B6 Q95R53 C4IY11 A0A1W4VZQ1 B4R378 B4Q1Q9 A0A2S2PG53 C4WY52 B4IK63 A0A1J1HHU7 B0XLY2 A0A182Q0S1 B3MQG1
A0A067QUM8 A0A1W4WMU8 N6T527 A0A088AQU4 A0A2A3EDB8 A0A1B6HLV1 A0A1B6F8X6 D6WEM3 A0A1Q3FS32 K7J2A5 A0A232F565 E2BVR3 A0A026X0W6 A0A195EZA2 A0A182GCV2 A0A1Y1MUQ1 A0A151WZF6 F4X2A3 E9IHT8 A0A151J6S3 A0A158P389 A0A154PE27 A0A023EKX8 A0A195CEV6 Q17HX8 A0A1I8NX44 B0XGR3 A0A1B6LP33 E0VTZ2 A0A1A9WCJ5 W8BSM6 A0A1I8MGX2 A0A1S4F1Y0 A0A182PM69 A0A0K8W233 A0A034W1U3 A0A0C9R2X2 A0A2M4DP35 A0A1A9XDV9 A0A0L0BRE4 A0A0M4EUT1 B4L1R2 A0A182V7F6 A0A0P4VLH9 T1HNH3 A0A182N0Y6 B4MEJ2 A0A084VIU5 A0A182XY43 A0A0V0GCT9 A0A1S3DGM5 A0A182RJ22 A0A182M8Y1 A0A182FM96 A0A182VVB8 A0A182J456 B4JM62 A0A3B0K5N9 A0A195BVF1 A0A182TQ91 A0A182X5B8 A0A0N0BDQ0 A0A1B0CVH5 A0A2S2RAM8 B4H3R8 W5JPG1 Q29JB5 A0A0A9WEG4 A0A182I9L8 A0A224XWG2 A0A182SR27 B3NU89 Q9VYU5 Q962B6 Q95R53 C4IY11 A0A1W4VZQ1 B4R378 B4Q1Q9 A0A2S2PG53 C4WY52 B4IK63 A0A1J1HHU7 B0XLY2 A0A182Q0S1 B3MQG1
Pubmed
19121390
26354079
22118469
24845553
23537049
18362917
+ More
19820115 20075255 28648823 20798317 24508170 26483478 28004739 21719571 21282665 21347285 24945155 17510324 20566863 24495485 25315136 25348373 26108605 17994087 27129103 24438588 25244985 20920257 23761445 15632085 25401762 26823975 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304
19820115 20075255 28648823 20798317 24508170 26483478 28004739 21719571 21282665 21347285 24945155 17510324 20566863 24495485 25315136 25348373 26108605 17994087 27129103 24438588 25244985 20920257 23761445 15632085 25401762 26823975 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304
EMBL
BABH01025900
ODYU01009323
SOQ53660.1
KQ460324
KPJ15845.1
AGBW02011024
+ More
OWR47353.1 NWSH01004146 PCG65429.1 KQ459249 KPJ02409.1 KK852992 KDR12765.1 APGK01043776 KB741017 KB631644 ENN75274.1 ERL84958.1 KZ288272 PBC29735.1 GECU01032034 JAS75672.1 GECZ01023215 JAS46554.1 KQ971326 EFA00870.1 GFDL01004802 JAV30243.1 AAZX01008245 NNAY01000914 OXU25944.1 GL450951 EFN80221.1 KK107037 EZA61950.1 KQ981905 KYN33635.1 JXUM01054857 JXUM01054858 JXUM01054859 KQ561840 KXJ77379.1 GEZM01020141 JAV89371.1 KQ982649 KYQ53147.1 GL888568 EGI59438.1 GL763294 EFZ19844.1 KQ979796 KYN19085.1 ADTU01007866 KQ434869 KZC09450.1 GAPW01003763 JAC09835.1 KQ977935 KYM98756.1 CH477244 EAT46299.1 DS233058 EDS27786.1 GEBQ01014524 JAT25453.1 DS235777 EEB16848.1 GAMC01006643 GAMC01006642 JAB99913.1 GDHF01007178 GDHF01002600 JAI45136.1 JAI49714.1 GAKP01010293 GAKP01010292 JAC48659.1 GBYB01010579 JAG80346.1 GGFL01015134 MBW79312.1 JRES01001467 KNC22650.1 CP012528 ALC48897.1 CH933810 EDW06715.2 GDKW01001328 JAI55267.1 ACPB03021429 CH940664 EDW62967.1 ATLV01013409 KE524855 KFB37889.1 GECL01000187 JAP05937.1 AXCM01007137 CH916371 EDV91823.1 OUUW01000003 SPP78778.1 KQ976403 KYM92240.1 KQ435859 KOX70498.1 AJWK01030574 AJWK01030575 GGMS01017846 MBY87049.1 CH479207 EDW31019.1 ADMH02000879 ETN64775.1 CH379063 EAL32386.2 GBHO01037466 GBHO01037465 GBHO01037464 GBRD01017153 GDHC01020358 GDHC01005264 GDHC01004085 JAG06138.1 JAG06139.1 JAG06140.1 JAG48674.1 JAP98270.1 JAQ13365.1 JAQ14544.1 APCN01001003 GFTR01003564 JAW12862.1 CH954180 EDV46004.1 AE014298 AAF48091.2 AAN09297.1 AHN59607.1 AHN59608.1 AY038185 AAK72697.1 AY061609 AAL29157.1 BT083442 ACR10174.1 CM000366 EDX17680.1 CM000162 EDX02484.2 GGMR01015788 MBY28407.1 ABLF02040099 ABLF02040119 AK343024 BAH72822.1 CH480851 EDW51435.1 CVRI01000004 CRK87467.1 DS234934 EDS35550.1 AXCN02001373 CH902621 EDV44587.1
OWR47353.1 NWSH01004146 PCG65429.1 KQ459249 KPJ02409.1 KK852992 KDR12765.1 APGK01043776 KB741017 KB631644 ENN75274.1 ERL84958.1 KZ288272 PBC29735.1 GECU01032034 JAS75672.1 GECZ01023215 JAS46554.1 KQ971326 EFA00870.1 GFDL01004802 JAV30243.1 AAZX01008245 NNAY01000914 OXU25944.1 GL450951 EFN80221.1 KK107037 EZA61950.1 KQ981905 KYN33635.1 JXUM01054857 JXUM01054858 JXUM01054859 KQ561840 KXJ77379.1 GEZM01020141 JAV89371.1 KQ982649 KYQ53147.1 GL888568 EGI59438.1 GL763294 EFZ19844.1 KQ979796 KYN19085.1 ADTU01007866 KQ434869 KZC09450.1 GAPW01003763 JAC09835.1 KQ977935 KYM98756.1 CH477244 EAT46299.1 DS233058 EDS27786.1 GEBQ01014524 JAT25453.1 DS235777 EEB16848.1 GAMC01006643 GAMC01006642 JAB99913.1 GDHF01007178 GDHF01002600 JAI45136.1 JAI49714.1 GAKP01010293 GAKP01010292 JAC48659.1 GBYB01010579 JAG80346.1 GGFL01015134 MBW79312.1 JRES01001467 KNC22650.1 CP012528 ALC48897.1 CH933810 EDW06715.2 GDKW01001328 JAI55267.1 ACPB03021429 CH940664 EDW62967.1 ATLV01013409 KE524855 KFB37889.1 GECL01000187 JAP05937.1 AXCM01007137 CH916371 EDV91823.1 OUUW01000003 SPP78778.1 KQ976403 KYM92240.1 KQ435859 KOX70498.1 AJWK01030574 AJWK01030575 GGMS01017846 MBY87049.1 CH479207 EDW31019.1 ADMH02000879 ETN64775.1 CH379063 EAL32386.2 GBHO01037466 GBHO01037465 GBHO01037464 GBRD01017153 GDHC01020358 GDHC01005264 GDHC01004085 JAG06138.1 JAG06139.1 JAG06140.1 JAG48674.1 JAP98270.1 JAQ13365.1 JAQ14544.1 APCN01001003 GFTR01003564 JAW12862.1 CH954180 EDV46004.1 AE014298 AAF48091.2 AAN09297.1 AHN59607.1 AHN59608.1 AY038185 AAK72697.1 AY061609 AAL29157.1 BT083442 ACR10174.1 CM000366 EDX17680.1 CM000162 EDX02484.2 GGMR01015788 MBY28407.1 ABLF02040099 ABLF02040119 AK343024 BAH72822.1 CH480851 EDW51435.1 CVRI01000004 CRK87467.1 DS234934 EDS35550.1 AXCN02001373 CH902621 EDV44587.1
Proteomes
UP000005204
UP000053240
UP000007151
UP000218220
UP000053268
UP000027135
+ More
UP000192223 UP000019118 UP000030742 UP000005203 UP000242457 UP000007266 UP000002358 UP000215335 UP000008237 UP000053097 UP000078541 UP000069940 UP000249989 UP000075809 UP000007755 UP000078492 UP000005205 UP000076502 UP000078542 UP000008820 UP000095300 UP000002320 UP000009046 UP000091820 UP000095301 UP000075885 UP000092443 UP000037069 UP000092553 UP000009192 UP000075903 UP000015103 UP000075884 UP000008792 UP000030765 UP000076408 UP000079169 UP000075900 UP000075883 UP000069272 UP000075920 UP000075880 UP000001070 UP000268350 UP000078540 UP000075902 UP000076407 UP000053105 UP000092461 UP000008744 UP000000673 UP000001819 UP000075840 UP000075901 UP000008711 UP000000803 UP000192221 UP000000304 UP000002282 UP000007819 UP000001292 UP000183832 UP000075886 UP000007801
UP000192223 UP000019118 UP000030742 UP000005203 UP000242457 UP000007266 UP000002358 UP000215335 UP000008237 UP000053097 UP000078541 UP000069940 UP000249989 UP000075809 UP000007755 UP000078492 UP000005205 UP000076502 UP000078542 UP000008820 UP000095300 UP000002320 UP000009046 UP000091820 UP000095301 UP000075885 UP000092443 UP000037069 UP000092553 UP000009192 UP000075903 UP000015103 UP000075884 UP000008792 UP000030765 UP000076408 UP000079169 UP000075900 UP000075883 UP000069272 UP000075920 UP000075880 UP000001070 UP000268350 UP000078540 UP000075902 UP000076407 UP000053105 UP000092461 UP000008744 UP000000673 UP000001819 UP000075840 UP000075901 UP000008711 UP000000803 UP000192221 UP000000304 UP000002282 UP000007819 UP000001292 UP000183832 UP000075886 UP000007801
Interpro
Gene 3D
ProteinModelPortal
H9J9V1
A0A2H1WKQ0
A0A0N1I8Q8
A0A212F0V5
A0A2A4J0M7
A0A194QAF1
+ More
A0A067QUM8 A0A1W4WMU8 N6T527 A0A088AQU4 A0A2A3EDB8 A0A1B6HLV1 A0A1B6F8X6 D6WEM3 A0A1Q3FS32 K7J2A5 A0A232F565 E2BVR3 A0A026X0W6 A0A195EZA2 A0A182GCV2 A0A1Y1MUQ1 A0A151WZF6 F4X2A3 E9IHT8 A0A151J6S3 A0A158P389 A0A154PE27 A0A023EKX8 A0A195CEV6 Q17HX8 A0A1I8NX44 B0XGR3 A0A1B6LP33 E0VTZ2 A0A1A9WCJ5 W8BSM6 A0A1I8MGX2 A0A1S4F1Y0 A0A182PM69 A0A0K8W233 A0A034W1U3 A0A0C9R2X2 A0A2M4DP35 A0A1A9XDV9 A0A0L0BRE4 A0A0M4EUT1 B4L1R2 A0A182V7F6 A0A0P4VLH9 T1HNH3 A0A182N0Y6 B4MEJ2 A0A084VIU5 A0A182XY43 A0A0V0GCT9 A0A1S3DGM5 A0A182RJ22 A0A182M8Y1 A0A182FM96 A0A182VVB8 A0A182J456 B4JM62 A0A3B0K5N9 A0A195BVF1 A0A182TQ91 A0A182X5B8 A0A0N0BDQ0 A0A1B0CVH5 A0A2S2RAM8 B4H3R8 W5JPG1 Q29JB5 A0A0A9WEG4 A0A182I9L8 A0A224XWG2 A0A182SR27 B3NU89 Q9VYU5 Q962B6 Q95R53 C4IY11 A0A1W4VZQ1 B4R378 B4Q1Q9 A0A2S2PG53 C4WY52 B4IK63 A0A1J1HHU7 B0XLY2 A0A182Q0S1 B3MQG1
A0A067QUM8 A0A1W4WMU8 N6T527 A0A088AQU4 A0A2A3EDB8 A0A1B6HLV1 A0A1B6F8X6 D6WEM3 A0A1Q3FS32 K7J2A5 A0A232F565 E2BVR3 A0A026X0W6 A0A195EZA2 A0A182GCV2 A0A1Y1MUQ1 A0A151WZF6 F4X2A3 E9IHT8 A0A151J6S3 A0A158P389 A0A154PE27 A0A023EKX8 A0A195CEV6 Q17HX8 A0A1I8NX44 B0XGR3 A0A1B6LP33 E0VTZ2 A0A1A9WCJ5 W8BSM6 A0A1I8MGX2 A0A1S4F1Y0 A0A182PM69 A0A0K8W233 A0A034W1U3 A0A0C9R2X2 A0A2M4DP35 A0A1A9XDV9 A0A0L0BRE4 A0A0M4EUT1 B4L1R2 A0A182V7F6 A0A0P4VLH9 T1HNH3 A0A182N0Y6 B4MEJ2 A0A084VIU5 A0A182XY43 A0A0V0GCT9 A0A1S3DGM5 A0A182RJ22 A0A182M8Y1 A0A182FM96 A0A182VVB8 A0A182J456 B4JM62 A0A3B0K5N9 A0A195BVF1 A0A182TQ91 A0A182X5B8 A0A0N0BDQ0 A0A1B0CVH5 A0A2S2RAM8 B4H3R8 W5JPG1 Q29JB5 A0A0A9WEG4 A0A182I9L8 A0A224XWG2 A0A182SR27 B3NU89 Q9VYU5 Q962B6 Q95R53 C4IY11 A0A1W4VZQ1 B4R378 B4Q1Q9 A0A2S2PG53 C4WY52 B4IK63 A0A1J1HHU7 B0XLY2 A0A182Q0S1 B3MQG1
Ontologies
Topology
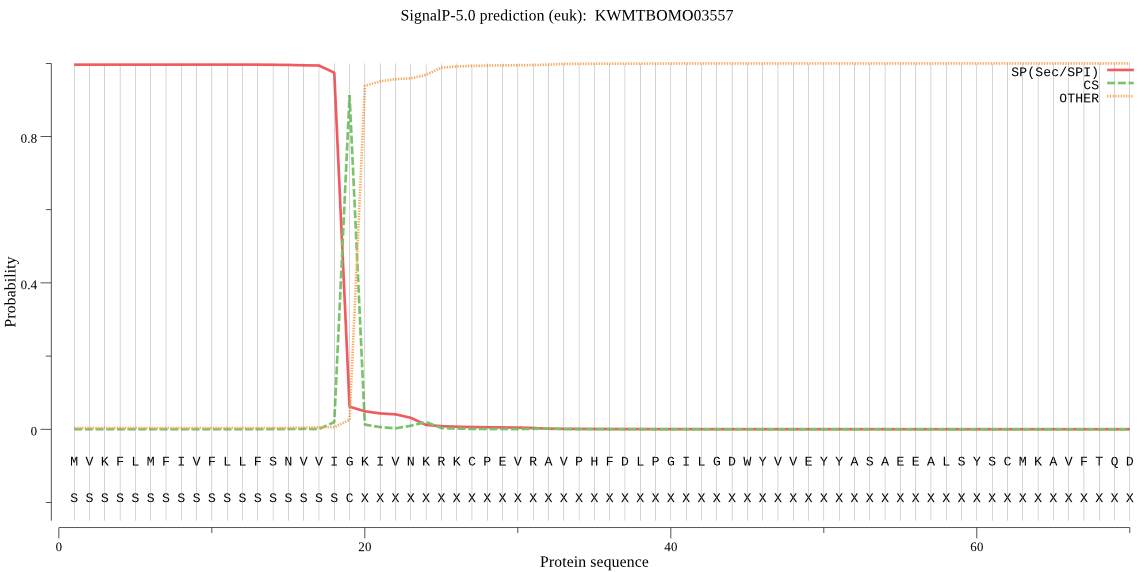
SignalP
Position: 1 - 19,
Likelihood: 0.996390
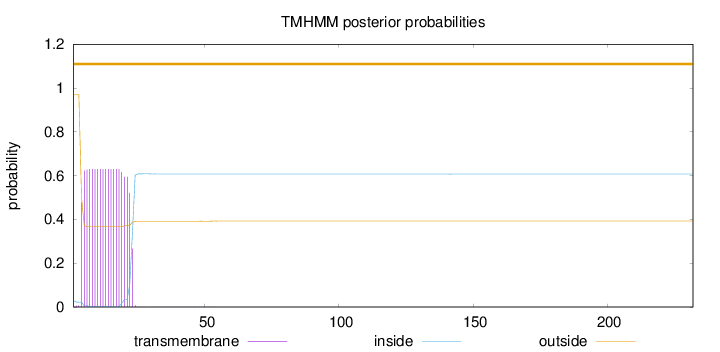
Length:
232
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
11.92925
Exp number, first 60 AAs:
11.92489
Total prob of N-in:
0.02898
POSSIBLE N-term signal
sequence
outside
1 - 232
Population Genetic Test Statistics
Pi
225.569895
Theta
163.857772
Tajima's D
1.191418
CLR
0.148178
CSRT
0.715014249287536
Interpretation
Uncertain
