Gene
KWMTBOMO03556
Pre Gene Modal
BGIBMGA010007
Annotation
putative_DNA_helicase_[Cotesia_vestalis_bracovirus]
Full name
ATP-dependent DNA helicase
Location in the cell
Mitochondrial Reliability : 1.233 PlasmaMembrane Reliability : 1.258
Sequence
CDS
ATGCCACGTTATTATACTTGGAATGCTTCATCGAAGAATTTTCAAAGACGGAAGCAAGGCGATGCGGTTCCTGGGTATCCAGATGTGCGTTCTACTGATGCTCTTGGTCGTATGTATACAGTTCATCCAAAGAATAATGAATGTTTCTATTTGCGGTTGTTGCTGGTAAATGTGCGTGGGCCAACGTCATTTGAGTCACTACGAACTGTTAATGGTGTAATATTCCCAACATATCGTGCTGCATGTGAAGAATTGAAGTTATTAGAAAACGATAGTCATTGGGATACGACAATCGCTGAAGCCATTATCTCTGCATCTCCAAGTCAGATACGCACATTATTCGCTATCATAATTTCGACATGTTTTCCATCAAACCCATGTAACCTGTGGCACAAATACAAGGATAATATGTCAGAAGATATTTTACATCAAATTCGTGTCAGTTCCAGAAATCACGATATTGAGATGAATGAGGAGATACATAATCGTGCTTTACTCTTGATCGAAGATATGTGTTACCTCATGTGCGGTAATTTATTAATCAGGTTAGGAATGCCAGCACCATATCGTGAAATGAATGACGCATTTAATCGAGAATTGGAAAGGGAACGTGAATATGATCACCAGGAATTAGATTTAGTAGTTCAAACGAATGTACCCCTGTTGAATTACCAACAAAAGGAAGTTTATGATACTTTAATGAAGGCAATCGATGATGAAAATGGTGGTTTATATTTCCTAGATGCCCCTGGTGGAACTGGCAAGACATTCCTCATGTCATTAGTTTTAGCAACTGTTCGGGCGAGATTTAACATAGCGGTTGCAGTTGCTTCTTCTGGAATAGCAGCCACATTGTTAGAAGGATGCCGTACGGCTCATTCAGCTTTCAAATTACCGTTAAATCTTCAAACTATTGAAGAACCAACGTGTAATATTGCAAAACACTCAGCAATGGCCAAAGTTTTAGCGGTATCGAAAATCATCATCTGGGACGAATGCACAATGGCGCATAAACGTACTTAA
Protein
MPRYYTWNASSKNFQRRKQGDAVPGYPDVRSTDALGRMYTVHPKNNECFYLRLLLVNVRGPTSFESLRTVNGVIFPTYRAACEELKLLENDSHWDTTIAEAIISASPSQIRTLFAIIISTCFPSNPCNLWHKYKDNMSEDILHQIRVSSRNHDIEMNEEIHNRALLLIEDMCYLMCGNLLIRLGMPAPYREMNDAFNRELEREREYDHQELDLVVQTNVPLLNYQQKEVYDTLMKAIDDENGGLYFLDAPGGTGKTFLMSLVLATVRARFNIAVAVASSGIAATLLEGCRTAHSAFKLPLNLQTIEEPTCNIAKHSAMAKVLAVSKIIIWDECTMAHKRT
Summary
Catalytic Activity
ATP + H2O = ADP + H(+) + phosphate
Cofactor
Mg(2+)
Similarity
Belongs to the helicase family.
Uniprot
A0A1L7NV02
S4PSG3
F4ZH17
H9JGG9
E2AZ86
A0A3S2LQP6
+ More
A0A0B7BQW2 A0A0B7BQV8 A0A3S2NR36 A0A0B7BTF0 X1X891 A0A0B7BRN8 X1WRV4 X1WU44 X1WQP7 A0A0A1WF35 A0A0C2BKN3 X1WUI1 A0A0C2G6A8 X1WPL7 A0A0C2G2C8 A0A0D6L4E4 A0A016W7D9 J9M4E0 A0A0C2DC56 A0A0C2CWG5 A0A0C2G6C2 A0A368FRT6 A0A368FI23 A0A368GPV9 A0A0C2GPE7 A0A016SC70 A0A0B7BRP2 A0A3S2NIC2 A0A0B7BU26 A0A087U8T2 A0A0D6LGL1 A0A0D6L8U7 A0A267GN76 A0A267GEW9 A0A267F5E2 A0A267GWL2 A0A267F4D8 A0A1B6FHF9 A0A368H513 A0A1S3DI71 A0A267F2R0 A0A182ERV7 X1WX02 X1X109 J9LFV8 A0A0C2DFS3 J9LSB3 A0A1S3T8U0 J9LB24 A0A267DHT9 A0A267EIS0 A0A267F6F6 A0A267FMY4 A0A267FG92 A0A267G6G0 A0A182EUU1 A0A267G464 A0A2C9JJ58 A0A267EUX6 A0A267F175 A0A267GBS1 A0A267GS15 A0A267ETE1 A0A267ELF5 A0A267GG99 A0A267GJF9 A0A267DCD1 A0A267DB74 A0A1I8JBS8 A0A2S2R387 A0A2S2QPJ4 A0A0L8HDK9 A0A267DIK3 A0A2H5SHM7 A0A267FCK1 A0A015KY09 A0A267ESS5 A0A2H5U846 A0A267H554 A0A267H341
A0A0B7BQW2 A0A0B7BQV8 A0A3S2NR36 A0A0B7BTF0 X1X891 A0A0B7BRN8 X1WRV4 X1WU44 X1WQP7 A0A0A1WF35 A0A0C2BKN3 X1WUI1 A0A0C2G6A8 X1WPL7 A0A0C2G2C8 A0A0D6L4E4 A0A016W7D9 J9M4E0 A0A0C2DC56 A0A0C2CWG5 A0A0C2G6C2 A0A368FRT6 A0A368FI23 A0A368GPV9 A0A0C2GPE7 A0A016SC70 A0A0B7BRP2 A0A3S2NIC2 A0A0B7BU26 A0A087U8T2 A0A0D6LGL1 A0A0D6L8U7 A0A267GN76 A0A267GEW9 A0A267F5E2 A0A267GWL2 A0A267F4D8 A0A1B6FHF9 A0A368H513 A0A1S3DI71 A0A267F2R0 A0A182ERV7 X1WX02 X1X109 J9LFV8 A0A0C2DFS3 J9LSB3 A0A1S3T8U0 J9LB24 A0A267DHT9 A0A267EIS0 A0A267F6F6 A0A267FMY4 A0A267FG92 A0A267G6G0 A0A182EUU1 A0A267G464 A0A2C9JJ58 A0A267EUX6 A0A267F175 A0A267GBS1 A0A267GS15 A0A267ETE1 A0A267ELF5 A0A267GG99 A0A267GJF9 A0A267DCD1 A0A267DB74 A0A1I8JBS8 A0A2S2R387 A0A2S2QPJ4 A0A0L8HDK9 A0A267DIK3 A0A2H5SHM7 A0A267FCK1 A0A015KY09 A0A267ESS5 A0A2H5U846 A0A267H554 A0A267H341
EC Number
3.6.4.12
Pubmed
EMBL
LC056060
BAW33744.1
GAIX01013838
JAA78722.1
HQ009558
AEE09607.1
+ More
BABH01039432 GL444143 EFN61253.1 RSAL01000340 RVE42412.1 HACG01048804 CEK95669.1 HACG01048799 CEK95664.1 RSAL01000338 RVE42429.1 HACG01048801 CEK95666.1 ABLF02039404 HACG01048798 CEK95663.1 ABLF02017703 ABLF02041883 ABLF02039418 ABLF02041882 ABLF02015085 ABLF02015086 GBXI01016780 JAC97511.1 KN778727 KIH44308.1 ABLF02016067 ABLF02055484 KN739098 KIH54434.1 ABLF02016932 KN747262 KIH51216.1 KE126587 EPB66060.1 JARK01000714 EYC35207.1 ABLF02042290 KN726708 KIH67418.1 KN739487 KIH54222.1 KN743176 KIH52626.1 JOJR01000723 RCN34924.1 JOJR01001507 RCN30639.1 JOJR01000111 RCN45090.1 KN730521 KIH60924.1 JARK01001590 EYB87952.1 HACG01048803 CEK95668.1 RSAL01001639 RVE40746.1 HACG01048800 CEK95665.1 KK118751 KFM73771.1 KE125929 EPB66767.1 KE125664 EPB67478.1 NIVC01000229 PAA87485.1 NIVC01000366 PAA84583.1 NIVC01001359 PAA68995.1 NIVC01000111 PAA90396.1 NIVC01001445 PAA67902.1 GECZ01020386 JAS49383.1 JOJR01000020 RCN50729.1 NIVC01001486 PAA67379.1 UYRW01006776 VDM94602.1 ABLF02041819 ABLF02020814 ABLF02049665 ABLF02012908 ABLF02042833 KN726282 KIH68828.1 ABLF02024220 ABLF02041348 ABLF02018127 NIVC01004230 PAA48122.1 NIVC01002034 PAA61410.1 NIVC01001383 PAA68602.1 NIVC01000900 PAA75136.1 NIVC01001115 PAA72069.1 NIVC01000566 PAA80832.1 UYRW01009134 VDM97499.1 PAA80831.1 NIVC01001660 PAA65355.1 NIVC01001478 PAA67511.1 NIVC01000419 PAA83453.1 NIVC01000175 PAA88803.1 NIVC01001722 PAA64736.1 NIVC01001945 PAA62370.1 NIVC01000381 PAA84309.1 NIVC01000291 PAA86185.1 NIVC01004666 PAA46875.1 NIVC01004811 PAA46541.1 GGMS01015273 MBY84476.1 GGMS01010390 MBY79593.1 KQ418414 KOF87426.1 NIVC01003989 PAA49045.1 BDIQ01000091 GBC29771.1 NIVC01001160 PAA71495.1 JEMT01022708 EXX64911.1 NIVC01002018 NIVC01001743 NIVC01000051 PAA61561.1 PAA64516.1 PAA92464.1 BDIQ01000188 GBC50999.1 NIVC01000042 PAA92669.1 NIVC01000040 PAA92731.1
BABH01039432 GL444143 EFN61253.1 RSAL01000340 RVE42412.1 HACG01048804 CEK95669.1 HACG01048799 CEK95664.1 RSAL01000338 RVE42429.1 HACG01048801 CEK95666.1 ABLF02039404 HACG01048798 CEK95663.1 ABLF02017703 ABLF02041883 ABLF02039418 ABLF02041882 ABLF02015085 ABLF02015086 GBXI01016780 JAC97511.1 KN778727 KIH44308.1 ABLF02016067 ABLF02055484 KN739098 KIH54434.1 ABLF02016932 KN747262 KIH51216.1 KE126587 EPB66060.1 JARK01000714 EYC35207.1 ABLF02042290 KN726708 KIH67418.1 KN739487 KIH54222.1 KN743176 KIH52626.1 JOJR01000723 RCN34924.1 JOJR01001507 RCN30639.1 JOJR01000111 RCN45090.1 KN730521 KIH60924.1 JARK01001590 EYB87952.1 HACG01048803 CEK95668.1 RSAL01001639 RVE40746.1 HACG01048800 CEK95665.1 KK118751 KFM73771.1 KE125929 EPB66767.1 KE125664 EPB67478.1 NIVC01000229 PAA87485.1 NIVC01000366 PAA84583.1 NIVC01001359 PAA68995.1 NIVC01000111 PAA90396.1 NIVC01001445 PAA67902.1 GECZ01020386 JAS49383.1 JOJR01000020 RCN50729.1 NIVC01001486 PAA67379.1 UYRW01006776 VDM94602.1 ABLF02041819 ABLF02020814 ABLF02049665 ABLF02012908 ABLF02042833 KN726282 KIH68828.1 ABLF02024220 ABLF02041348 ABLF02018127 NIVC01004230 PAA48122.1 NIVC01002034 PAA61410.1 NIVC01001383 PAA68602.1 NIVC01000900 PAA75136.1 NIVC01001115 PAA72069.1 NIVC01000566 PAA80832.1 UYRW01009134 VDM97499.1 PAA80831.1 NIVC01001660 PAA65355.1 NIVC01001478 PAA67511.1 NIVC01000419 PAA83453.1 NIVC01000175 PAA88803.1 NIVC01001722 PAA64736.1 NIVC01001945 PAA62370.1 NIVC01000381 PAA84309.1 NIVC01000291 PAA86185.1 NIVC01004666 PAA46875.1 NIVC01004811 PAA46541.1 GGMS01015273 MBY84476.1 GGMS01010390 MBY79593.1 KQ418414 KOF87426.1 NIVC01003989 PAA49045.1 BDIQ01000091 GBC29771.1 NIVC01001160 PAA71495.1 JEMT01022708 EXX64911.1 NIVC01002018 NIVC01001743 NIVC01000051 PAA61561.1 PAA64516.1 PAA92464.1 BDIQ01000188 GBC50999.1 NIVC01000042 PAA92669.1 NIVC01000040 PAA92731.1
Proteomes
PRIDE
Interpro
ProteinModelPortal
A0A1L7NV02
S4PSG3
F4ZH17
H9JGG9
E2AZ86
A0A3S2LQP6
+ More
A0A0B7BQW2 A0A0B7BQV8 A0A3S2NR36 A0A0B7BTF0 X1X891 A0A0B7BRN8 X1WRV4 X1WU44 X1WQP7 A0A0A1WF35 A0A0C2BKN3 X1WUI1 A0A0C2G6A8 X1WPL7 A0A0C2G2C8 A0A0D6L4E4 A0A016W7D9 J9M4E0 A0A0C2DC56 A0A0C2CWG5 A0A0C2G6C2 A0A368FRT6 A0A368FI23 A0A368GPV9 A0A0C2GPE7 A0A016SC70 A0A0B7BRP2 A0A3S2NIC2 A0A0B7BU26 A0A087U8T2 A0A0D6LGL1 A0A0D6L8U7 A0A267GN76 A0A267GEW9 A0A267F5E2 A0A267GWL2 A0A267F4D8 A0A1B6FHF9 A0A368H513 A0A1S3DI71 A0A267F2R0 A0A182ERV7 X1WX02 X1X109 J9LFV8 A0A0C2DFS3 J9LSB3 A0A1S3T8U0 J9LB24 A0A267DHT9 A0A267EIS0 A0A267F6F6 A0A267FMY4 A0A267FG92 A0A267G6G0 A0A182EUU1 A0A267G464 A0A2C9JJ58 A0A267EUX6 A0A267F175 A0A267GBS1 A0A267GS15 A0A267ETE1 A0A267ELF5 A0A267GG99 A0A267GJF9 A0A267DCD1 A0A267DB74 A0A1I8JBS8 A0A2S2R387 A0A2S2QPJ4 A0A0L8HDK9 A0A267DIK3 A0A2H5SHM7 A0A267FCK1 A0A015KY09 A0A267ESS5 A0A2H5U846 A0A267H554 A0A267H341
A0A0B7BQW2 A0A0B7BQV8 A0A3S2NR36 A0A0B7BTF0 X1X891 A0A0B7BRN8 X1WRV4 X1WU44 X1WQP7 A0A0A1WF35 A0A0C2BKN3 X1WUI1 A0A0C2G6A8 X1WPL7 A0A0C2G2C8 A0A0D6L4E4 A0A016W7D9 J9M4E0 A0A0C2DC56 A0A0C2CWG5 A0A0C2G6C2 A0A368FRT6 A0A368FI23 A0A368GPV9 A0A0C2GPE7 A0A016SC70 A0A0B7BRP2 A0A3S2NIC2 A0A0B7BU26 A0A087U8T2 A0A0D6LGL1 A0A0D6L8U7 A0A267GN76 A0A267GEW9 A0A267F5E2 A0A267GWL2 A0A267F4D8 A0A1B6FHF9 A0A368H513 A0A1S3DI71 A0A267F2R0 A0A182ERV7 X1WX02 X1X109 J9LFV8 A0A0C2DFS3 J9LSB3 A0A1S3T8U0 J9LB24 A0A267DHT9 A0A267EIS0 A0A267F6F6 A0A267FMY4 A0A267FG92 A0A267G6G0 A0A182EUU1 A0A267G464 A0A2C9JJ58 A0A267EUX6 A0A267F175 A0A267GBS1 A0A267GS15 A0A267ETE1 A0A267ELF5 A0A267GG99 A0A267GJF9 A0A267DCD1 A0A267DB74 A0A1I8JBS8 A0A2S2R387 A0A2S2QPJ4 A0A0L8HDK9 A0A267DIK3 A0A2H5SHM7 A0A267FCK1 A0A015KY09 A0A267ESS5 A0A2H5U846 A0A267H554 A0A267H341
Ontologies
GO
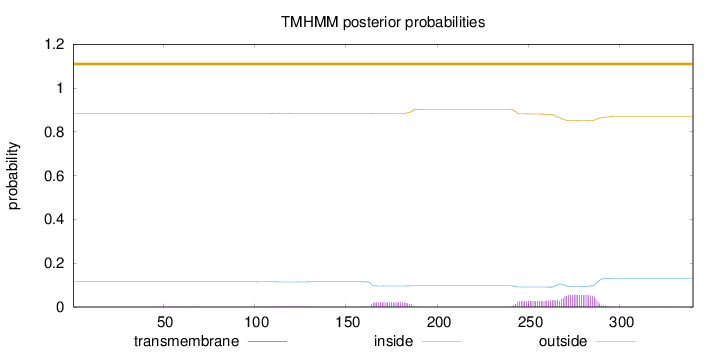
Topology
Length:
340
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.29289
Exp number, first 60 AAs:
0.00352
Total prob of N-in:
0.11537
outside
1 - 340
Population Genetic Test Statistics
Pi
176.761771
Theta
147.079784
Tajima's D
3.743567
CLR
0
CSRT
0.997650117494125
Interpretation
Possibly Balancing Selection
