Pre Gene Modal
BGIBMGA006292
Annotation
PREDICTED:_transcription-associated_zinc_ribbon_protein_isoform_X1_[Bombyx_mori]
Full name
DNA-directed RNA polymerase subunit
Location in the cell
Extracellular Reliability : 3.515
Sequence
CDS
ATGTCCACTGAAAGCCCGTTATCAACTAGTACAGCGTTTTGTGCTCGCTGTGGGTCCATATTACCCTTGCTACAAGAGTTTGGCTCCGTGAAATGCTACACTTGTAAAGCACATTATGAGGCTGATAATTATAGTAAAATGAAATTTCAATACACAATACATTTCAATACTATATCTGTGATCACAAATGAGAATATACTACATACTGATGGTCCTGAGGGTCCAGTTGTGGAGAGAAAATGCGCTAAATGCGGTTACGACAGAATGTCATATGCAACTCTACAGTTACGATCGGCCGATGAAGGACAAACAGTTTTCTACACATGTATAAAGTGCAAATACAAGGAAACTGAAAACTCCTAA
Protein
MSTESPLSTSTAFCARCGSILPLLQEFGSVKCYTCKAHYEADNYSKMKFQYTIHFNTISVITNENILHTDGPEGPVVERKCAKCGYDRMSYATLQLRSADEGQTVFYTCIKCKYKETENS
Summary
Description
DNA-dependent RNA polymerase catalyzes the transcription of DNA into RNA using the four ribonucleoside triphosphates as substrates.
Similarity
Belongs to the archaeal rpoM/eukaryotic RPA12/RPB9/RPC11 RNA polymerase family.
Uniprot
Q1ZYJ6
A0A2H1X1X9
A0A2A4JLT4
A0A212F0V8
A0A0N0PD94
A0A194QAA8
+ More
A0A1E1W007 S4PSG7 A0A0L7LNR5 U5EQG9 A0A0T6AVL2 A0A0L0BXV6 A0A1W4VPS8 D6WBZ4 B4LWP3 A0A2P8YV40 Q9VD81 A0A2A3E768 A0A1L8DID1 A0A1S3DL81 A0A158NPQ1 A0A1B0CLC9 A0A1I8P607 E2C5M5 A0A3B0JKV5 A0A2M4AVZ5 B4HM44 B4K8D8 B4QZ91 A0A067R9I3 A0A1Q3F0P4 A0A1I8MNL4 B4PKV3 Q16SW6 B4JU97 Q16SW7 B3NZY6 W8CDU1 B4NHN6 A0A034WQP8 A0A1Y1MPW4 B3LVZ2 A0A1B6GXN3 Q29AR6 B4G606 A0A182FA85 A0A2M4C3S9 W5J1F5 A0A182US58 A0A182XIB1 A0A182LFE1 Q7PR45 A0A182HT96 A0A0K8WIW5 A0A182UF37 A0A2R7WFG3 A0A0A9Z5P1 B0WS60 A0A2M4CYM7 A0A182K3C4 A0A182QE25 A0A151IMU2 A0A182RYG2 A0A0N7Z8V9 R4G7X7 A0A336LXJ7 A0A182H0M9 A0A232FMC8 K7IXE8 A0A2M3ZJW8 A0A2M3ZD73 E0VBV6 A0A195EL19 A0A182MLP1 A0A195AT83 A0A3L8DAA5 A0A0V0GDY6 A0A023EI75 A0A0M8ZN31 A0A0L7R308 A0A069DP51 A0A182W2W8 A0A1B6LDY9 A0A1A9VIX5 A0A1W4WYX6 E2B0G0 J9JU77 A0A224XPQ2 N6UG87 A0A182IKI2 A0A2M3ZD77 A0A182Y779 E9GG94 A0A2S2P0X7 A0A0P6GYA4 F4W7I7 A0A195ESW4 A0A0M5J7M9 A0A0P6BV12 A0A154P238
A0A1E1W007 S4PSG7 A0A0L7LNR5 U5EQG9 A0A0T6AVL2 A0A0L0BXV6 A0A1W4VPS8 D6WBZ4 B4LWP3 A0A2P8YV40 Q9VD81 A0A2A3E768 A0A1L8DID1 A0A1S3DL81 A0A158NPQ1 A0A1B0CLC9 A0A1I8P607 E2C5M5 A0A3B0JKV5 A0A2M4AVZ5 B4HM44 B4K8D8 B4QZ91 A0A067R9I3 A0A1Q3F0P4 A0A1I8MNL4 B4PKV3 Q16SW6 B4JU97 Q16SW7 B3NZY6 W8CDU1 B4NHN6 A0A034WQP8 A0A1Y1MPW4 B3LVZ2 A0A1B6GXN3 Q29AR6 B4G606 A0A182FA85 A0A2M4C3S9 W5J1F5 A0A182US58 A0A182XIB1 A0A182LFE1 Q7PR45 A0A182HT96 A0A0K8WIW5 A0A182UF37 A0A2R7WFG3 A0A0A9Z5P1 B0WS60 A0A2M4CYM7 A0A182K3C4 A0A182QE25 A0A151IMU2 A0A182RYG2 A0A0N7Z8V9 R4G7X7 A0A336LXJ7 A0A182H0M9 A0A232FMC8 K7IXE8 A0A2M3ZJW8 A0A2M3ZD73 E0VBV6 A0A195EL19 A0A182MLP1 A0A195AT83 A0A3L8DAA5 A0A0V0GDY6 A0A023EI75 A0A0M8ZN31 A0A0L7R308 A0A069DP51 A0A182W2W8 A0A1B6LDY9 A0A1A9VIX5 A0A1W4WYX6 E2B0G0 J9JU77 A0A224XPQ2 N6UG87 A0A182IKI2 A0A2M3ZD77 A0A182Y779 E9GG94 A0A2S2P0X7 A0A0P6GYA4 F4W7I7 A0A195ESW4 A0A0M5J7M9 A0A0P6BV12 A0A154P238
Pubmed
19121390
22118469
26354079
23622113
26227816
26108605
+ More
18362917 19820115 17994087 29403074 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 21347285 20798317 24845553 25315136 17550304 17510324 18057021 24495485 25348373 28004739 15632085 20920257 23761445 20966253 12364791 14747013 17210077 25401762 26823975 27129103 26483478 28648823 20075255 20566863 30249741 24945155 26334808 23537049 25244985 21292972 21719571
18362917 19820115 17994087 29403074 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 21347285 20798317 24845553 25315136 17550304 17510324 18057021 24495485 25348373 28004739 15632085 20920257 23761445 20966253 12364791 14747013 17210077 25401762 26823975 27129103 26483478 28648823 20075255 20566863 30249741 24945155 26334808 23537049 25244985 21292972 21719571
EMBL
BABH01025883
DQ432055
ABD76573.1
ODYU01012776
SOQ59248.1
NWSH01001070
+ More
PCG72779.1 AGBW02011024 OWR47357.1 KQ460324 KPJ15849.1 KQ459249 KPJ02412.1 GDQN01010737 JAT80317.1 GAIX01013818 JAA78742.1 JTDY01000477 KOB77004.1 GANO01003268 JAB56603.1 LJIG01022753 KRT78901.1 JRES01001168 KNC24857.1 KQ971309 EEZ99110.1 CH940650 EDW67708.1 PYGN01000343 PSN48110.1 AE014297 BT050591 AAF55918.1 ACJ73151.1 KZ288351 PBC27344.1 GFDF01007949 JAV06135.1 ADTU01022705 AJWK01017076 GL452776 EFN76736.1 OUUW01000007 SPP82957.1 GGFK01011646 MBW44967.1 CH480815 EDW43092.1 CH933806 EDW16520.1 CM000364 EDX13827.1 KK852840 KDR15193.1 GFDL01013919 JAV21126.1 CM000160 EDW97902.1 CH477664 EAT37552.1 CH916374 EDV91067.1 EAT37553.1 CH954462 CH954181 EDV45162.1 EDV49984.1 GAMC01001356 JAC05200.1 CH964272 EDW84646.1 GAKP01002874 JAC56078.1 GEZM01028055 GEZM01028054 GEZM01028053 JAV86505.1 CH902617 EDV41525.1 GECZ01002584 JAS67185.1 CM000070 EAL27283.1 CH479179 EDW23773.1 GGFJ01010784 MBW59925.1 ADMH02002183 ETN57997.1 AAAB01008859 EAA07851.5 APCN01000424 GDHF01001216 JAI51098.1 KK854672 PTY17790.1 GBHO01003790 GBHO01000547 GBRD01001211 GDHC01021243 JAG39814.1 JAG43057.1 JAG64610.1 JAP97385.1 DS232065 EDS33683.1 GGFL01006201 MBW70379.1 AXCN02000218 KQ977026 KYN06251.1 GDKW01002259 JAI54336.1 ACPB03008859 GAHY01001952 JAA75558.1 UFQT01000283 SSX22762.1 JXUM01101629 KQ564663 KXJ71939.1 NNAY01000020 OXU31884.1 AAZX01000007 GGFM01008070 MBW28821.1 GGFM01005756 MBW26507.1 DS235042 EEB10862.1 KQ978730 KYN28950.1 AXCM01000066 KQ976745 KYM75433.1 QOIP01000011 RLU17072.1 GECL01000070 JAP06054.1 GAPW01004983 JAC08615.1 KQ436126 KOX67437.1 KQ414663 KOC65218.1 GBGD01003512 JAC85377.1 GEBQ01018243 JAT21734.1 GL444608 EFN60802.1 ABLF02034733 GFTR01001960 JAW14466.1 APGK01021787 KB740364 KB632275 ENN80775.1 ERL91146.1 GGFM01005711 MBW26462.1 GL732543 EFX81535.1 GGMR01010456 MBY23075.1 GDIQ01026923 LRGB01000725 JAN67814.1 KZS16307.1 GL887844 EGI69860.1 KQ981986 KYN31246.1 CP012526 ALC47818.1 GDIP01009386 JAM94329.1 KQ434803 KZC05892.1
PCG72779.1 AGBW02011024 OWR47357.1 KQ460324 KPJ15849.1 KQ459249 KPJ02412.1 GDQN01010737 JAT80317.1 GAIX01013818 JAA78742.1 JTDY01000477 KOB77004.1 GANO01003268 JAB56603.1 LJIG01022753 KRT78901.1 JRES01001168 KNC24857.1 KQ971309 EEZ99110.1 CH940650 EDW67708.1 PYGN01000343 PSN48110.1 AE014297 BT050591 AAF55918.1 ACJ73151.1 KZ288351 PBC27344.1 GFDF01007949 JAV06135.1 ADTU01022705 AJWK01017076 GL452776 EFN76736.1 OUUW01000007 SPP82957.1 GGFK01011646 MBW44967.1 CH480815 EDW43092.1 CH933806 EDW16520.1 CM000364 EDX13827.1 KK852840 KDR15193.1 GFDL01013919 JAV21126.1 CM000160 EDW97902.1 CH477664 EAT37552.1 CH916374 EDV91067.1 EAT37553.1 CH954462 CH954181 EDV45162.1 EDV49984.1 GAMC01001356 JAC05200.1 CH964272 EDW84646.1 GAKP01002874 JAC56078.1 GEZM01028055 GEZM01028054 GEZM01028053 JAV86505.1 CH902617 EDV41525.1 GECZ01002584 JAS67185.1 CM000070 EAL27283.1 CH479179 EDW23773.1 GGFJ01010784 MBW59925.1 ADMH02002183 ETN57997.1 AAAB01008859 EAA07851.5 APCN01000424 GDHF01001216 JAI51098.1 KK854672 PTY17790.1 GBHO01003790 GBHO01000547 GBRD01001211 GDHC01021243 JAG39814.1 JAG43057.1 JAG64610.1 JAP97385.1 DS232065 EDS33683.1 GGFL01006201 MBW70379.1 AXCN02000218 KQ977026 KYN06251.1 GDKW01002259 JAI54336.1 ACPB03008859 GAHY01001952 JAA75558.1 UFQT01000283 SSX22762.1 JXUM01101629 KQ564663 KXJ71939.1 NNAY01000020 OXU31884.1 AAZX01000007 GGFM01008070 MBW28821.1 GGFM01005756 MBW26507.1 DS235042 EEB10862.1 KQ978730 KYN28950.1 AXCM01000066 KQ976745 KYM75433.1 QOIP01000011 RLU17072.1 GECL01000070 JAP06054.1 GAPW01004983 JAC08615.1 KQ436126 KOX67437.1 KQ414663 KOC65218.1 GBGD01003512 JAC85377.1 GEBQ01018243 JAT21734.1 GL444608 EFN60802.1 ABLF02034733 GFTR01001960 JAW14466.1 APGK01021787 KB740364 KB632275 ENN80775.1 ERL91146.1 GGFM01005711 MBW26462.1 GL732543 EFX81535.1 GGMR01010456 MBY23075.1 GDIQ01026923 LRGB01000725 JAN67814.1 KZS16307.1 GL887844 EGI69860.1 KQ981986 KYN31246.1 CP012526 ALC47818.1 GDIP01009386 JAM94329.1 KQ434803 KZC05892.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000053268
UP000037510
+ More
UP000037069 UP000192221 UP000007266 UP000008792 UP000245037 UP000000803 UP000242457 UP000079169 UP000005205 UP000092461 UP000095300 UP000008237 UP000268350 UP000001292 UP000009192 UP000000304 UP000027135 UP000095301 UP000002282 UP000008820 UP000001070 UP000008711 UP000007798 UP000007801 UP000001819 UP000008744 UP000069272 UP000000673 UP000075903 UP000076407 UP000075882 UP000007062 UP000075840 UP000075902 UP000002320 UP000075881 UP000075886 UP000078542 UP000075900 UP000015103 UP000069940 UP000249989 UP000215335 UP000002358 UP000009046 UP000078492 UP000075883 UP000078540 UP000279307 UP000053105 UP000053825 UP000075920 UP000078200 UP000192223 UP000000311 UP000007819 UP000019118 UP000030742 UP000075880 UP000076408 UP000000305 UP000076858 UP000007755 UP000078541 UP000092553 UP000076502
UP000037069 UP000192221 UP000007266 UP000008792 UP000245037 UP000000803 UP000242457 UP000079169 UP000005205 UP000092461 UP000095300 UP000008237 UP000268350 UP000001292 UP000009192 UP000000304 UP000027135 UP000095301 UP000002282 UP000008820 UP000001070 UP000008711 UP000007798 UP000007801 UP000001819 UP000008744 UP000069272 UP000000673 UP000075903 UP000076407 UP000075882 UP000007062 UP000075840 UP000075902 UP000002320 UP000075881 UP000075886 UP000078542 UP000075900 UP000015103 UP000069940 UP000249989 UP000215335 UP000002358 UP000009046 UP000078492 UP000075883 UP000078540 UP000279307 UP000053105 UP000053825 UP000075920 UP000078200 UP000192223 UP000000311 UP000007819 UP000019118 UP000030742 UP000075880 UP000076408 UP000000305 UP000076858 UP000007755 UP000078541 UP000092553 UP000076502
Pfam
PF01096 TFIIS_C
Interpro
ProteinModelPortal
Q1ZYJ6
A0A2H1X1X9
A0A2A4JLT4
A0A212F0V8
A0A0N0PD94
A0A194QAA8
+ More
A0A1E1W007 S4PSG7 A0A0L7LNR5 U5EQG9 A0A0T6AVL2 A0A0L0BXV6 A0A1W4VPS8 D6WBZ4 B4LWP3 A0A2P8YV40 Q9VD81 A0A2A3E768 A0A1L8DID1 A0A1S3DL81 A0A158NPQ1 A0A1B0CLC9 A0A1I8P607 E2C5M5 A0A3B0JKV5 A0A2M4AVZ5 B4HM44 B4K8D8 B4QZ91 A0A067R9I3 A0A1Q3F0P4 A0A1I8MNL4 B4PKV3 Q16SW6 B4JU97 Q16SW7 B3NZY6 W8CDU1 B4NHN6 A0A034WQP8 A0A1Y1MPW4 B3LVZ2 A0A1B6GXN3 Q29AR6 B4G606 A0A182FA85 A0A2M4C3S9 W5J1F5 A0A182US58 A0A182XIB1 A0A182LFE1 Q7PR45 A0A182HT96 A0A0K8WIW5 A0A182UF37 A0A2R7WFG3 A0A0A9Z5P1 B0WS60 A0A2M4CYM7 A0A182K3C4 A0A182QE25 A0A151IMU2 A0A182RYG2 A0A0N7Z8V9 R4G7X7 A0A336LXJ7 A0A182H0M9 A0A232FMC8 K7IXE8 A0A2M3ZJW8 A0A2M3ZD73 E0VBV6 A0A195EL19 A0A182MLP1 A0A195AT83 A0A3L8DAA5 A0A0V0GDY6 A0A023EI75 A0A0M8ZN31 A0A0L7R308 A0A069DP51 A0A182W2W8 A0A1B6LDY9 A0A1A9VIX5 A0A1W4WYX6 E2B0G0 J9JU77 A0A224XPQ2 N6UG87 A0A182IKI2 A0A2M3ZD77 A0A182Y779 E9GG94 A0A2S2P0X7 A0A0P6GYA4 F4W7I7 A0A195ESW4 A0A0M5J7M9 A0A0P6BV12 A0A154P238
A0A1E1W007 S4PSG7 A0A0L7LNR5 U5EQG9 A0A0T6AVL2 A0A0L0BXV6 A0A1W4VPS8 D6WBZ4 B4LWP3 A0A2P8YV40 Q9VD81 A0A2A3E768 A0A1L8DID1 A0A1S3DL81 A0A158NPQ1 A0A1B0CLC9 A0A1I8P607 E2C5M5 A0A3B0JKV5 A0A2M4AVZ5 B4HM44 B4K8D8 B4QZ91 A0A067R9I3 A0A1Q3F0P4 A0A1I8MNL4 B4PKV3 Q16SW6 B4JU97 Q16SW7 B3NZY6 W8CDU1 B4NHN6 A0A034WQP8 A0A1Y1MPW4 B3LVZ2 A0A1B6GXN3 Q29AR6 B4G606 A0A182FA85 A0A2M4C3S9 W5J1F5 A0A182US58 A0A182XIB1 A0A182LFE1 Q7PR45 A0A182HT96 A0A0K8WIW5 A0A182UF37 A0A2R7WFG3 A0A0A9Z5P1 B0WS60 A0A2M4CYM7 A0A182K3C4 A0A182QE25 A0A151IMU2 A0A182RYG2 A0A0N7Z8V9 R4G7X7 A0A336LXJ7 A0A182H0M9 A0A232FMC8 K7IXE8 A0A2M3ZJW8 A0A2M3ZD73 E0VBV6 A0A195EL19 A0A182MLP1 A0A195AT83 A0A3L8DAA5 A0A0V0GDY6 A0A023EI75 A0A0M8ZN31 A0A0L7R308 A0A069DP51 A0A182W2W8 A0A1B6LDY9 A0A1A9VIX5 A0A1W4WYX6 E2B0G0 J9JU77 A0A224XPQ2 N6UG87 A0A182IKI2 A0A2M3ZD77 A0A182Y779 E9GG94 A0A2S2P0X7 A0A0P6GYA4 F4W7I7 A0A195ESW4 A0A0M5J7M9 A0A0P6BV12 A0A154P238
PDB
6HLS
E-value=4.23833e-12,
Score=165
Ontologies
GO
Topology
Subcellular location
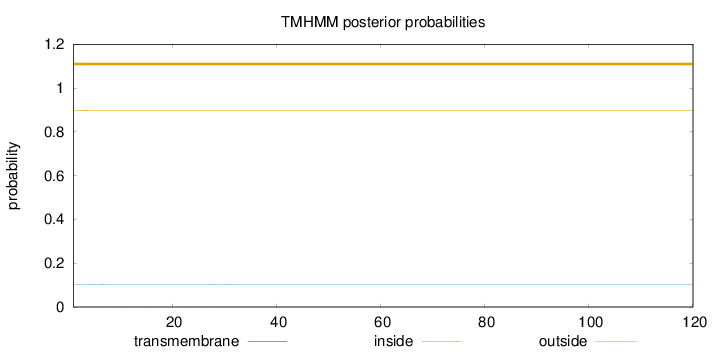
Length:
120
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02181
Exp number, first 60 AAs:
0.02106
Total prob of N-in:
0.10222
outside
1 - 120
Population Genetic Test Statistics
Pi
224.270474
Theta
206.167034
Tajima's D
0.245306
CLR
0
CSRT
0.437178141092945
Interpretation
Uncertain
