Gene
KWMTBOMO03549
Pre Gene Modal
BGIBMGA006291
Annotation
eclosion_hormone_[synthetic_construct]
Full name
Eclosion hormone
Alternative Name
EH
Ecdysis activator
Ecdysis activator
Location in the cell
Extracellular Reliability : 2.675
Sequence
CDS
ATGGCAAACAAACTGACCGCAGTCATCGTTGTTGCTCTCGCCGTGGCATTCATGGTCAACCTGGACTATGCCAACTGCAGCCCTGCCATCGCATCCAGCTACGACGCAATGGAAATTTGCATCGAGAACTGCGCGCAATGCAAAAAAATGTTCGGCCCTTGGTTCGAGGGATCCCTTTGCGCGGAGTCCTGCATCAAAGCCAGAGGCAAAGACATTCCGGAATGCGAAAGCTTCGCATCCATATCTCCATTCCTGAATAAGCTTTAA
Protein
MANKLTAVIVVALAVAFMVNLDYANCSPAIASSYDAMEICIENCAQCKKMFGPWFEGSLCAESCIKARGKDIPECESFASISPFLNKL
Summary
Description
Neuropeptide that triggers the performance of ecdysis behaviors at the end of a molt. It triggers adult behavior patterns: larval, pupal and adult ecdysis, and plasticization during the molt.
Similarity
Belongs to the insect eclosion hormone family.
Keywords
Complete proteome
Direct protein sequencing
Disulfide bond
Hormone
Neuropeptide
Reference proteome
Secreted
Signal
Feature
chain Eclosion hormone
Uniprot
P25331
P11919
Q5PXP1
A0A2A4J7N1
A0A2H1X1M5
Q0PYK0
+ More
A0A0S1U1V2 I4DL09 A0A0N0PDJ9 A0A194QAF5 A0A212F0S8 A0A0N1I9J1 A0A194QHZ4 A0A194QNW8 A0A194QI35 Q16QN2 A0A182K9H5 A0A1S4FSX6 A0A1B0GQM6 A0A182QPH7 A0A084VZL2 A0A1I8JV18 A0A182HXA0 B0X456 W5JM68 A0A182RBC3 A0A182W3T4 A0A182X862 A0NCV3 T1E8L0 A0A0N1PH64 A0A182NDL7 A0A336M1M9 A0A182YIP4 A0A336MWM7 A0A182FM43 Q1DGP2 A0A182H5W6 E0W1X8 T1JCY5 A0A0M4ELR6 A0A084VZL3 A0A023ECI0 A0A023EDT1 Q174X1 A0A023EE17 Q29A86 B4G2M2 B4QVQ2 B4IBH2 A0A0B4KGU5 B4PVB7 Q07892 A0A3B0K6N5 B0WXK2 A0A1W4URN1 B3P3D5 B3LVQ6
A0A0S1U1V2 I4DL09 A0A0N0PDJ9 A0A194QAF5 A0A212F0S8 A0A0N1I9J1 A0A194QHZ4 A0A194QNW8 A0A194QI35 Q16QN2 A0A182K9H5 A0A1S4FSX6 A0A1B0GQM6 A0A182QPH7 A0A084VZL2 A0A1I8JV18 A0A182HXA0 B0X456 W5JM68 A0A182RBC3 A0A182W3T4 A0A182X862 A0NCV3 T1E8L0 A0A0N1PH64 A0A182NDL7 A0A336M1M9 A0A182YIP4 A0A336MWM7 A0A182FM43 Q1DGP2 A0A182H5W6 E0W1X8 T1JCY5 A0A0M4ELR6 A0A084VZL3 A0A023ECI0 A0A023EDT1 Q174X1 A0A023EE17 Q29A86 B4G2M2 B4QVQ2 B4IBH2 A0A0B4KGU5 B4PVB7 Q07892 A0A3B0K6N5 B0WXK2 A0A1W4URN1 B3P3D5 B3LVQ6
Pubmed
EMBL
D10135
M27808
M26922
AY822476
AAV69026.1
NWSH01002602
+ More
PCG67879.1 ODYU01012776 SOQ59250.1 DQ668369 ABG66962.1 KT005968 ALM30319.1 AK401977 BAM18599.1 KQ460324 KPJ15850.1 KQ459249 KPJ02414.1 AGBW02011024 OWR47359.1 KQ460078 KPJ17851.1 KQ458761 KPJ05178.1 KPJ05176.1 KPJ05177.1 CH477741 EAT36711.1 AJVK01007397 AXCN02001751 ATLV01018857 KE525251 KFB43406.1 APCN01005163 DS232329 EDS40133.1 ADMH02000723 ETN65246.1 AAAB01008849 EAU77164.1 GAMD01002673 JAA98917.1 KPJ17853.1 UFQS01000247 UFQT01000247 SSX01980.1 SSX22357.1 UFQS01002094 UFQT01002094 SSX13218.1 SSX32657.1 CH901007 EAT32321.1 JXUM01026757 KQ560766 KXJ80967.1 DS235873 EEB19572.1 JH432085 CP012526 ALC46290.1 KFB43407.1 GAPW01006555 JAC07043.1 GAPW01006662 JAC06936.1 CH477404 EAT41662.1 JXUM01020902 GAPW01006552 KQ560585 JAC07046.1 KXJ81787.1 CM000070 EAL27464.1 CH479179 EDW24067.1 CM000364 EDX12565.1 CH480827 EDW44730.1 AE014297 AGB96048.1 CM000160 EDW95786.1 X72302 X72303 BT023233 OUUW01000005 SPP80651.1 DS232168 EDS36532.1 CH954181 EDV48715.1 CH902617 EDV43680.2
PCG67879.1 ODYU01012776 SOQ59250.1 DQ668369 ABG66962.1 KT005968 ALM30319.1 AK401977 BAM18599.1 KQ460324 KPJ15850.1 KQ459249 KPJ02414.1 AGBW02011024 OWR47359.1 KQ460078 KPJ17851.1 KQ458761 KPJ05178.1 KPJ05176.1 KPJ05177.1 CH477741 EAT36711.1 AJVK01007397 AXCN02001751 ATLV01018857 KE525251 KFB43406.1 APCN01005163 DS232329 EDS40133.1 ADMH02000723 ETN65246.1 AAAB01008849 EAU77164.1 GAMD01002673 JAA98917.1 KPJ17853.1 UFQS01000247 UFQT01000247 SSX01980.1 SSX22357.1 UFQS01002094 UFQT01002094 SSX13218.1 SSX32657.1 CH901007 EAT32321.1 JXUM01026757 KQ560766 KXJ80967.1 DS235873 EEB19572.1 JH432085 CP012526 ALC46290.1 KFB43407.1 GAPW01006555 JAC07043.1 GAPW01006662 JAC06936.1 CH477404 EAT41662.1 JXUM01020902 GAPW01006552 KQ560585 JAC07046.1 KXJ81787.1 CM000070 EAL27464.1 CH479179 EDW24067.1 CM000364 EDX12565.1 CH480827 EDW44730.1 AE014297 AGB96048.1 CM000160 EDW95786.1 X72302 X72303 BT023233 OUUW01000005 SPP80651.1 DS232168 EDS36532.1 CH954181 EDV48715.1 CH902617 EDV43680.2
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000008820
+ More
UP000075881 UP000092462 UP000075886 UP000030765 UP000075902 UP000075840 UP000002320 UP000000673 UP000075900 UP000075920 UP000076407 UP000007062 UP000075884 UP000076408 UP000069272 UP000069940 UP000249989 UP000009046 UP000092553 UP000001819 UP000008744 UP000000304 UP000001292 UP000000803 UP000002282 UP000268350 UP000192221 UP000008711 UP000007801
UP000075881 UP000092462 UP000075886 UP000030765 UP000075902 UP000075840 UP000002320 UP000000673 UP000075900 UP000075920 UP000076407 UP000007062 UP000075884 UP000076408 UP000069272 UP000069940 UP000249989 UP000009046 UP000092553 UP000001819 UP000008744 UP000000304 UP000001292 UP000000803 UP000002282 UP000268350 UP000192221 UP000008711 UP000007801
Pfam
PF04736 Eclosion
ProteinModelPortal
P25331
P11919
Q5PXP1
A0A2A4J7N1
A0A2H1X1M5
Q0PYK0
+ More
A0A0S1U1V2 I4DL09 A0A0N0PDJ9 A0A194QAF5 A0A212F0S8 A0A0N1I9J1 A0A194QHZ4 A0A194QNW8 A0A194QI35 Q16QN2 A0A182K9H5 A0A1S4FSX6 A0A1B0GQM6 A0A182QPH7 A0A084VZL2 A0A1I8JV18 A0A182HXA0 B0X456 W5JM68 A0A182RBC3 A0A182W3T4 A0A182X862 A0NCV3 T1E8L0 A0A0N1PH64 A0A182NDL7 A0A336M1M9 A0A182YIP4 A0A336MWM7 A0A182FM43 Q1DGP2 A0A182H5W6 E0W1X8 T1JCY5 A0A0M4ELR6 A0A084VZL3 A0A023ECI0 A0A023EDT1 Q174X1 A0A023EE17 Q29A86 B4G2M2 B4QVQ2 B4IBH2 A0A0B4KGU5 B4PVB7 Q07892 A0A3B0K6N5 B0WXK2 A0A1W4URN1 B3P3D5 B3LVQ6
A0A0S1U1V2 I4DL09 A0A0N0PDJ9 A0A194QAF5 A0A212F0S8 A0A0N1I9J1 A0A194QHZ4 A0A194QNW8 A0A194QI35 Q16QN2 A0A182K9H5 A0A1S4FSX6 A0A1B0GQM6 A0A182QPH7 A0A084VZL2 A0A1I8JV18 A0A182HXA0 B0X456 W5JM68 A0A182RBC3 A0A182W3T4 A0A182X862 A0NCV3 T1E8L0 A0A0N1PH64 A0A182NDL7 A0A336M1M9 A0A182YIP4 A0A336MWM7 A0A182FM43 Q1DGP2 A0A182H5W6 E0W1X8 T1JCY5 A0A0M4ELR6 A0A084VZL3 A0A023ECI0 A0A023EDT1 Q174X1 A0A023EE17 Q29A86 B4G2M2 B4QVQ2 B4IBH2 A0A0B4KGU5 B4PVB7 Q07892 A0A3B0K6N5 B0WXK2 A0A1W4URN1 B3P3D5 B3LVQ6
Ontologies
GO
PANTHER
Topology
Subcellular location
Secreted
SignalP
Position: 1 - 26,
Likelihood: 0.994430
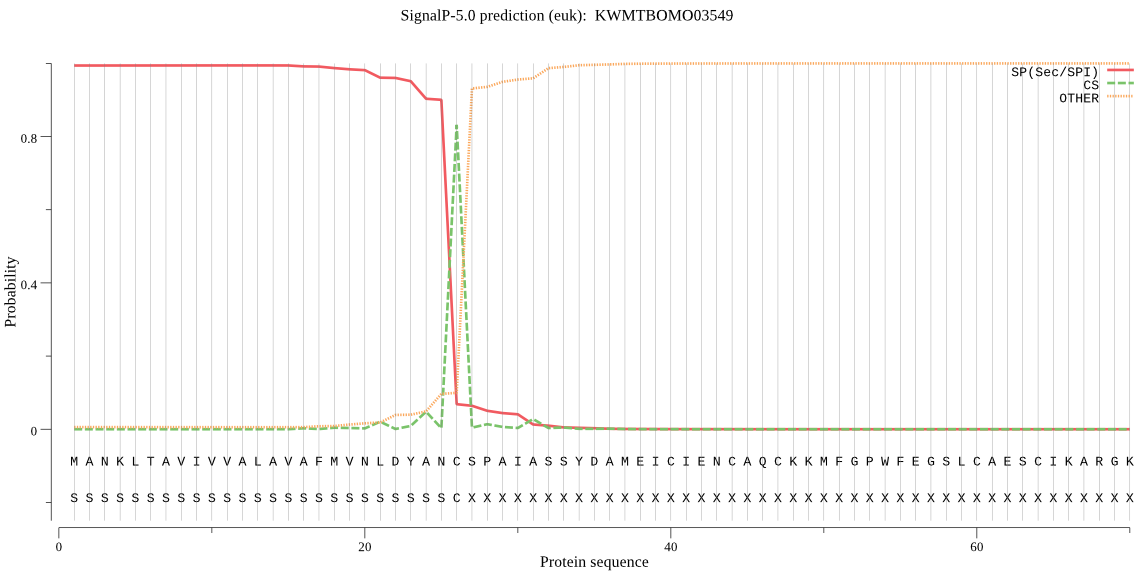
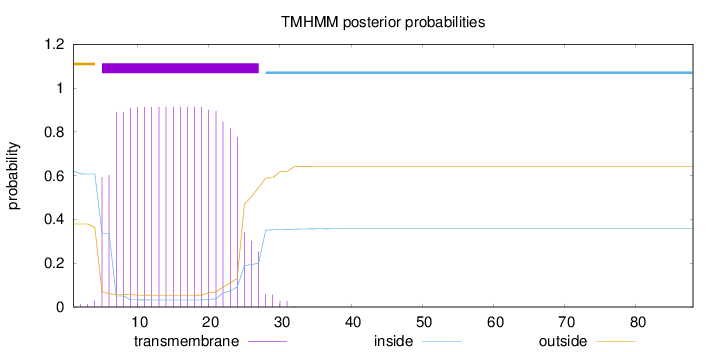
Length:
88
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
18.3994
Exp number, first 60 AAs:
18.39939
Total prob of N-in:
0.62068
POSSIBLE N-term signal
sequence
outside
1 - 4
TMhelix
5 - 27
inside
28 - 88
Population Genetic Test Statistics
Pi
191.76287
Theta
184.785183
Tajima's D
-1.098651
CLR
1.874884
CSRT
0.123793810309485
Interpretation
Uncertain
