Gene
KWMTBOMO03536 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA006251
Annotation
PREDICTED:_zonadhesin-like_isoform_X1_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 2.698
Sequence
CDS
ATGAAATGTCCGGTGCCCGCCAAATGTAGACCTACTTGCTCTGTGCCGAATCCACCGAACTGTCCACCTTACAAGCCGCCTACTGAGAACGAGGACGGATGTCGATGTCAACCAGGGTACATCTTGTCTGAAACAGAAGGGAAATGTGTTAAAATAGAAACCTGCCCTACTGGTCAGTGTCCGGTGCCCGCCAAATGTAGACCTACTTGCTCTGTGCCGAATCCACCGAACTGTCCACCTTACAAGCAGCCTACTGAGAACGAGGACGGATGTCGATGTCAACCAGGGTACATCTTGTCTGAAACAGAAGGGAAATGTGTTAAAATAGAAACCTGCCCTACTGGTCAGTGTCCGGTGCCCGCCAAATGTAGACCTACTTGCTCTGTGCCGAATCCACCGAACTGTCCACCTTACAAGCAGCCTACTGAGAACGAGGACGGATGTCGATGTCAACCAGGGTACATCTTGTCTGAAACAGAAGGGAAATGTGTTAAAATAGAAACCTGCCCTAGTTCGTGTTCTGGTAACGAAACGCGCGTATACTGCAAGGCTGGCTGTCCCACTGACTATTGTCCAACAAGCGACAGCCGCGCTGTAATTGCTTGCGATCCCCCCTACCCATGTAATCCGGGCTGCGTTTGTAAACCAAATTATCGACGCCTGAGTCAGGATGACGAGCGATGTGTGCTGACCACAGACTGTCCTCCAGTGAAGTGCACACGACCGAATGAAGTTTGGAATCCTTGTACACAATGTTTGGCGGAACGATGCGAGGATGCCAACAGTCCCTCTAACCTGAATTGTCCTCCTCAGCTGCGCCCTTATTGCAATCCGCAGTGCGTGTGTGCAAACGGTTATTACAGGAACAACAATGACGTTTGTGTCCCGAAGAAAGCTTGTCCTGGTCAGTGTCCGGTGCCCGCCAGGTGGAGACCTACCTGCTCTGTGCCGAATCCACCGAATTGTCCACCTTACAAGCCGCCTGCTGAGAACAAGGACGGATGTCAATGTCAAGAAGGGTACATCTTGTCTGAAGCAGAAGGAAAATGTATTAAAATAGAAGAATGTCCTAGAACATCAGGCTGTAATGGAGATCCACATGCAAAAGTGACGAAGTGTCCGTTACCTTGCCCGTCCACTTGCAAATCACCAGAGGCTATAGGTTGTAAGAAAAGGTGCGATCCGGTTGGATGTGAATGCGAGTATGGCTACATACTATCAGATATAGGAGGAAAATGTATTAAGCCTGAAGAATGCCCAGGCGGAAACCCGTGTGGATCAAATGGTACATTTGTTAAATGCAAATCAAGGTGTCCGAATGACTATTGTCCTGTCAACGATAATCGTGGAGTTATAATTTGTGAAGGCTTTCCATTAGCCCCCTGTTACTCTGGATGTATTTGTAAGCTAAATTATAAAAGACGCAGCGAGACAGACCGAAGATGCATTCTGTCTACAGATTGCCCTCCTGTAAATTGCACTAGACCCAACGAAGTCTGGTGCCCGTCACCACCGCCCTGTTTACAAGAAACCTGCGATGATGTCGACCTTCCTCCGGTACCTTGTAACGAATTTATAAAAAGCAGTCCTAGATGCATCTGCAAAGATAATTACTTCAGGAACGATAGCGGAATCTGCGTTTGCGCTCAGGAATGTCGCGACATAAACGCGCCTGATTATAATCGCAAGAAGCGTGATATATTCATATGCTCCGCAACCGCGCACAATACAACTGAAGGTCCGTGCAATGCAAACGAAACGCTCGTATCCTGCAAAGCTGGCTGTCCCACCGACTATTGTCCAACAAGCGACAGCCGCGCTGTAGTTGCTTGCGATCCCCCCTACCCATGTAATCCGGGCTGCGTTTGTAAACCAAATTATAAGCTCTTGAGTCAGGATGACGAGCGATGTGTGCTAACCACAGACTGTCCTCCTGTAAATTGCACTAGACCCAATGAAGTCTGGAGCCCGTCACCACCGCCCTGTTTACAAGAAACCTGCGATGATGTCGACGTTACTCCGGTACCTTGTAACGAGTTTATACAAAGCAGCCCTAGATGCATCTGCAAAGATAATTACTTCAGGAACGATAGCGGAATCTGCGTTTCCGCTCAGGAATGTCGCGACATAAACGCGCGTGATTATAATCGCAAGAAGCGTGATATACCGGAATGTGGTTATAACGAAGTACTGTCGGATTCCAAAATAGTGTGTCCCCCTCAGACTTGTGAGAGCATATACACTACGTATTTATGTGAGTCTAGGGGTGATGAAGTTGGATGTAACTGTTTATCTGGATATTTGAGGAACAGCAGTGGTATATGCATTCCCAGTGAGGAATGTTTTCAAGCGGAGAATTGCTCGCTTCCTTCTGAATGTATACCGACATGCGCAATTCCGAATCCCAATTGTTCCTACGTTCAGGAAGTTTCCTTCAAAAGAGAAGCCTGTTATTGTAAGCCTGGTTTCATATTATCCGAGGTCAATGGGGAATGCATTAAGATCGAAAATTGTCCAAAACAACAATGCGGGGTAAACGGAACATATTCCGATTGCGGCTTTCGATGCCCAAACCAGTATTGTCCTGAAGACGACAGTTTACTCCAAATAGCGTGTAAGCCTGGGCGACCCTGCCCGCCTGGCTGCGTATGTAAGGATAACTACAAAAGAAAAAGTCGCGAAGAAGACGATGTTTGCGTACTCGCCTCAGATTGTCCACCCGTCAATTGCACTAGGCCTAATGAAGTTTGGAATCCCTGTTCGCAATGTTTGGCGGAACGATGCGAAGATGCTAACACTACTAATTGTTATGATCCGCCCGGCTCAAACTGTGACCCTAGGTGCGTGTGTGTAGATGGGTATAAAAGAAACAAAGATGACGTTTGCGTCCCGGAGAAAGCGTGTCCTGGTCGATGTCCGATACCGATTAAATGTAGACCGACATGTGCTGTTCCTAATCCAAAAAAATGTTCTAAGTATAAATTACCTGCCAACAATGAGGATGGGTGTCAATGTAAGAAAGGATACATTTTGTCGGAACTCGATGGGAAATGTGTTGCAATCGACAAATGTCCCAAAATGGGAGGTTGCAACGGTGATCCGCATGCGGTAATAAAAGAATGTCCGTTGGCTTGTCCGTCTACTTGTGAGGCACCGGAGGCTAAGCCCTGTAAAAAGAAATGTGCTCCTGTCGGATGCGAATGCGAATCTGGTTATATACTTTCGAAAACAAAAGGCAAATGTATCAGCCCCGAAGAATGCCCAGGCTAG
Protein
MKCPVPAKCRPTCSVPNPPNCPPYKPPTENEDGCRCQPGYILSETEGKCVKIETCPTGQCPVPAKCRPTCSVPNPPNCPPYKQPTENEDGCRCQPGYILSETEGKCVKIETCPTGQCPVPAKCRPTCSVPNPPNCPPYKQPTENEDGCRCQPGYILSETEGKCVKIETCPSSCSGNETRVYCKAGCPTDYCPTSDSRAVIACDPPYPCNPGCVCKPNYRRLSQDDERCVLTTDCPPVKCTRPNEVWNPCTQCLAERCEDANSPSNLNCPPQLRPYCNPQCVCANGYYRNNNDVCVPKKACPGQCPVPARWRPTCSVPNPPNCPPYKPPAENKDGCQCQEGYILSEAEGKCIKIEECPRTSGCNGDPHAKVTKCPLPCPSTCKSPEAIGCKKRCDPVGCECEYGYILSDIGGKCIKPEECPGGNPCGSNGTFVKCKSRCPNDYCPVNDNRGVIICEGFPLAPCYSGCICKLNYKRRSETDRRCILSTDCPPVNCTRPNEVWCPSPPPCLQETCDDVDLPPVPCNEFIKSSPRCICKDNYFRNDSGICVCAQECRDINAPDYNRKKRDIFICSATAHNTTEGPCNANETLVSCKAGCPTDYCPTSDSRAVVACDPPYPCNPGCVCKPNYKLLSQDDERCVLTTDCPPVNCTRPNEVWSPSPPPCLQETCDDVDVTPVPCNEFIQSSPRCICKDNYFRNDSGICVSAQECRDINARDYNRKKRDIPECGYNEVLSDSKIVCPPQTCESIYTTYLCESRGDEVGCNCLSGYLRNSSGICIPSEECFQAENCSLPSECIPTCAIPNPNCSYVQEVSFKREACYCKPGFILSEVNGECIKIENCPKQQCGVNGTYSDCGFRCPNQYCPEDDSLLQIACKPGRPCPPGCVCKDNYKRKSREEDDVCVLASDCPPVNCTRPNEVWNPCSQCLAERCEDANTTNCYDPPGSNCDPRCVCVDGYKRNKDDVCVPEKACPGRCPIPIKCRPTCAVPNPKKCSKYKLPANNEDGCQCKKGYILSELDGKCVAIDKCPKMGGCNGDPHAVIKECPLACPSTCEAPEAKPCKKKCAPVGCECESGYILSKTKGKCISPEECPG
Summary
Similarity
Belongs to the serpin family.
Uniprot
A0A2A4J071
A0A194QAG0
A0A2H1VKZ4
A0A194QQM3
A0A2W1B8Z9
A0A2A4J7B9
+ More
A0A2W1BQG2 A0A2A4IXH8 A0A087UJV8 A0A212EGT4 A0A0K0FVD9 A0A2A4IW13 A0A2H1VQQ3 A0A0L7L697 A0A1B0D4C2 A0A224YHH5 A0A131YWN5 L7LQD3 L7LR33 A0A1B0CYV2 A0A1B0D8S3 A0A2L2XYN6 A0A090N0V2 A0A1B0CYV0 A0A0L7L220 A0A1B0CET6 A0A1B0GMI5 A0A1B0GHG9 A0A368GXV9 W2ST73
A0A2W1BQG2 A0A2A4IXH8 A0A087UJV8 A0A212EGT4 A0A0K0FVD9 A0A2A4IW13 A0A2H1VQQ3 A0A0L7L697 A0A1B0D4C2 A0A224YHH5 A0A131YWN5 L7LQD3 L7LR33 A0A1B0CYV2 A0A1B0D8S3 A0A2L2XYN6 A0A090N0V2 A0A1B0CYV0 A0A0L7L220 A0A1B0CET6 A0A1B0GMI5 A0A1B0GHG9 A0A368GXV9 W2ST73
EMBL
NWSH01004181
PCG65391.1
KQ459249
KPJ02419.1
ODYU01002951
SOQ41102.1
+ More
KQ461181 KPJ07649.1 KZ150221 PZC72102.1 NWSH01002542 PCG68017.1 KZ149981 PZC75815.1 NWSH01005913 PCG63850.1 KK120157 KFM77647.1 AGBW02015025 OWR40695.1 PCG63849.1 ODYU01003852 SOQ43127.1 JTDY01002637 KOB71038.1 AJVK01024288 GFPF01005269 MAA16415.1 GEDV01005971 JAP82586.1 GACK01010873 JAA54161.1 GACK01010874 JAA54160.1 AJVK01020418 AJVK01027694 IAAA01002394 LAA00998.1 LN609530 CEF71333.1 JTDY01003492 KOB69475.1 AJWK01009195 AJWK01009196 AJVK01011483 AJWK01004621 JOJR01000037 RCN49202.1 KI663210 ETN72713.1
KQ461181 KPJ07649.1 KZ150221 PZC72102.1 NWSH01002542 PCG68017.1 KZ149981 PZC75815.1 NWSH01005913 PCG63850.1 KK120157 KFM77647.1 AGBW02015025 OWR40695.1 PCG63849.1 ODYU01003852 SOQ43127.1 JTDY01002637 KOB71038.1 AJVK01024288 GFPF01005269 MAA16415.1 GEDV01005971 JAP82586.1 GACK01010873 JAA54161.1 GACK01010874 JAA54160.1 AJVK01020418 AJVK01027694 IAAA01002394 LAA00998.1 LN609530 CEF71333.1 JTDY01003492 KOB69475.1 AJWK01009195 AJWK01009196 AJVK01011483 AJWK01004621 JOJR01000037 RCN49202.1 KI663210 ETN72713.1
Proteomes
Interpro
Gene 3D
ProteinModelPortal
A0A2A4J071
A0A194QAG0
A0A2H1VKZ4
A0A194QQM3
A0A2W1B8Z9
A0A2A4J7B9
+ More
A0A2W1BQG2 A0A2A4IXH8 A0A087UJV8 A0A212EGT4 A0A0K0FVD9 A0A2A4IW13 A0A2H1VQQ3 A0A0L7L697 A0A1B0D4C2 A0A224YHH5 A0A131YWN5 L7LQD3 L7LR33 A0A1B0CYV2 A0A1B0D8S3 A0A2L2XYN6 A0A090N0V2 A0A1B0CYV0 A0A0L7L220 A0A1B0CET6 A0A1B0GMI5 A0A1B0GHG9 A0A368GXV9 W2ST73
A0A2W1BQG2 A0A2A4IXH8 A0A087UJV8 A0A212EGT4 A0A0K0FVD9 A0A2A4IW13 A0A2H1VQQ3 A0A0L7L697 A0A1B0D4C2 A0A224YHH5 A0A131YWN5 L7LQD3 L7LR33 A0A1B0CYV2 A0A1B0D8S3 A0A2L2XYN6 A0A090N0V2 A0A1B0CYV0 A0A0L7L220 A0A1B0CET6 A0A1B0GMI5 A0A1B0GHG9 A0A368GXV9 W2ST73
Ontologies
GO
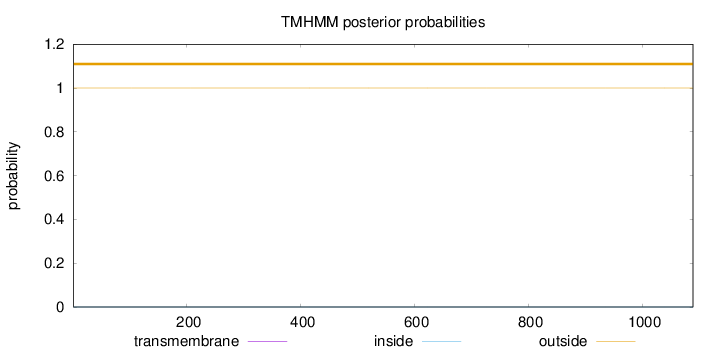
Topology
Length:
1089
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00001
outside
1 - 1089
Population Genetic Test Statistics
Pi
212.498845
Theta
175.630191
Tajima's D
0.561912
CLR
0.43243
CSRT
0.530673466326684
Interpretation
Uncertain
