Gene
KWMTBOMO03535 Validated by peptides from experiments
Annotation
PREDICTED:_zonadhesin-like_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 3.527
Sequence
CDS
ATGTGTGCTGACCACAGACTGTCTTGTCAGTGGAAACCTTTTTCAGCTCCAGTCACGTGCACACGACCGAATGAAGTTTGGAATCCGTGTGCACAATGTTTGGCGGAACGATGCGAGGATCTCAACAGTACCTCTAACCAGAATTGTCCTGCTGAACTGCGTTCCTACTGCAGGCCGCAGTGCGTTTGTGCAAACGGCTATTACAGGAACAACAATGACGTTTGTGTCCCGAAGAAAGCTTGTCCTGGTCAGTGTCCGGTGCCCGCCAGGTGTAGACCTACCTGCTCTGTGCCGAATCCACCGAACTGTCCACCTTACAAGCCGCCTACTGAGAACGAGGACGGATGTCGATGTCAACCAGGGTACATCTTGTCTGAAACAGAAGGGAAATGTGTTAAAATAGAAACCTGCCCTAGTCCGTGCAATGCAAACGAAACGCTCGTGTACTGCAAAGCTGGCTGTCCCACCGACTATTGTCCAACAAGCGACAGCCGCGCTATAGTTGCTTGCTCACCCCCCTATCCATGTAATCCGGGCTGCGTTTGTAAACCAAATTATCGAATCTTAAGTCAGGATGACAAGCGATGTGTGCTGACCACAGACTGTCGTAAGTACAACATAGAGTATAGTAAAAATAGTTTTATTGGGCATTACGGTGATATCTTCTGGTTGTCGTAG
Protein
MCADHRLSCQWKPFSAPVTCTRPNEVWNPCAQCLAERCEDLNSTSNQNCPAELRSYCRPQCVCANGYYRNNNDVCVPKKACPGQCPVPARCRPTCSVPNPPNCPPYKPPTENEDGCRCQPGYILSETEGKCVKIETCPSPCNANETLVYCKAGCPTDYCPTSDSRAIVACSPPYPCNPGCVCKPNYRILSQDDKRCVLTTDCRKYNIEYSKNSFIGHYGDIFWLS
Summary
Similarity
Belongs to the serpin family.
Uniprot
A0A2A4J071
A0A2H1VKZ4
A0A0L7LWK3
A0A2W1BA04
A0A0V0Z791
A0A2A4J6N9
+ More
A0A0V0Z7M5 A0A2A4IW13 A0A2A4IXH8 A0A0V0Z760 A0A0V1CXK4 A0A0V0Z7C1 A0A0V0S5K9 A0A0V0Z793 A0A1B0CE59 A0A0V0VIZ3 A0A0V0U5N5 A0A0V1LSL0 A0A0V0S563 E5S2I0 A0A0V1NT84 A0A0V0S515 A0A0V0U5E1 A0A0V1B9Z3 A0A0V1B9Y2 A0A2P6L3Q8 A0A1B0CYU8 A0A1J1IUR1 A0A2A4J7B9 A0A2A4J556 A0A0V0WZ71 A0A0V0WZ93 A0A0V1EPP1 A0A0V1EPN2 A0A2W1BQG2 A0A0V1J3G8 A0A0V1EPP9 H9J5D7 A0A0V1FGY8 A0A2H1VAX4 A0A0V1J3E7 A0A3G1T1R3 A0A0V1MZF6 Q8I4B9 Q9U1T5 A0A0L7L697 A0A1J1HI61 A0A0V1H1U7 H9JMY6 A0A087UJV8 A0A0K0EAF5 A0A1J1HLL5 A0A2Z6FLF1 H2L2K9 A0A0L7L220 A0A182PDJ4 A0A224YFL8
A0A0V0Z7M5 A0A2A4IW13 A0A2A4IXH8 A0A0V0Z760 A0A0V1CXK4 A0A0V0Z7C1 A0A0V0S5K9 A0A0V0Z793 A0A1B0CE59 A0A0V0VIZ3 A0A0V0U5N5 A0A0V1LSL0 A0A0V0S563 E5S2I0 A0A0V1NT84 A0A0V0S515 A0A0V0U5E1 A0A0V1B9Z3 A0A0V1B9Y2 A0A2P6L3Q8 A0A1B0CYU8 A0A1J1IUR1 A0A2A4J7B9 A0A2A4J556 A0A0V0WZ71 A0A0V0WZ93 A0A0V1EPP1 A0A0V1EPN2 A0A2W1BQG2 A0A0V1J3G8 A0A0V1EPP9 H9J5D7 A0A0V1FGY8 A0A2H1VAX4 A0A0V1J3E7 A0A3G1T1R3 A0A0V1MZF6 Q8I4B9 Q9U1T5 A0A0L7L697 A0A1J1HI61 A0A0V1H1U7 H9JMY6 A0A087UJV8 A0A0K0EAF5 A0A1J1HLL5 A0A2Z6FLF1 H2L2K9 A0A0L7L220 A0A182PDJ4 A0A224YFL8
EMBL
NWSH01004181
PCG65391.1
ODYU01002951
SOQ41102.1
JTDY01000004
KOB79511.1
+ More
KZ150198 PZC72302.1 JYDQ01000343 KRY08319.1 NWSH01003013 PCG67110.1 KRY08323.1 NWSH01005913 PCG63849.1 PCG63850.1 KRY08320.1 JYDI01000077 KRY53944.1 KRY08317.1 JYDL01000037 KRX21786.1 KRY08321.1 AJWK01008601 JYDN01000032 KRX63333.1 JYDJ01000057 KRX46475.1 JYDW01000009 KRZ62443.1 KRX21783.1 JYDM01000104 KRZ87210.1 KRX21785.1 KRX46473.1 JYDH01000076 KRY33812.1 KRY33814.1 MWRG01001996 PRD33209.1 AJVK01020418 CVRI01000059 CRL03442.1 NWSH01002542 PCG68017.1 PCG67111.1 JYDK01000035 KRX81073.1 KRX81072.1 JYDR01000016 KRY75722.1 KRY75723.1 KZ149981 PZC75815.1 JYDS01000045 KRZ29499.1 KRY75718.1 BABH01039220 BABH01039221 BABH01039222 BABH01039223 JYDT01000094 KRY85257.1 ODYU01001563 SOQ37951.1 KRZ29498.1 MG770321 AXY94923.1 JYDO01000022 KRZ77159.1 BX284605 CAD56616.1 CAB54473.2 JTDY01002637 KOB71038.1 CVRI01000004 CRK87701.1 JYDP01000166 KRZ04345.1 BABH01029067 KK120157 KFM77647.1 CVRI01000010 CRK88925.1 LC384844 BBE28987.1 CCE71937.1 JTDY01003492 KOB69475.1 GFPF01005340 MAA16486.1
KZ150198 PZC72302.1 JYDQ01000343 KRY08319.1 NWSH01003013 PCG67110.1 KRY08323.1 NWSH01005913 PCG63849.1 PCG63850.1 KRY08320.1 JYDI01000077 KRY53944.1 KRY08317.1 JYDL01000037 KRX21786.1 KRY08321.1 AJWK01008601 JYDN01000032 KRX63333.1 JYDJ01000057 KRX46475.1 JYDW01000009 KRZ62443.1 KRX21783.1 JYDM01000104 KRZ87210.1 KRX21785.1 KRX46473.1 JYDH01000076 KRY33812.1 KRY33814.1 MWRG01001996 PRD33209.1 AJVK01020418 CVRI01000059 CRL03442.1 NWSH01002542 PCG68017.1 PCG67111.1 JYDK01000035 KRX81073.1 KRX81072.1 JYDR01000016 KRY75722.1 KRY75723.1 KZ149981 PZC75815.1 JYDS01000045 KRZ29499.1 KRY75718.1 BABH01039220 BABH01039221 BABH01039222 BABH01039223 JYDT01000094 KRY85257.1 ODYU01001563 SOQ37951.1 KRZ29498.1 MG770321 AXY94923.1 JYDO01000022 KRZ77159.1 BX284605 CAD56616.1 CAB54473.2 JTDY01002637 KOB71038.1 CVRI01000004 CRK87701.1 JYDP01000166 KRZ04345.1 BABH01029067 KK120157 KFM77647.1 CVRI01000010 CRK88925.1 LC384844 BBE28987.1 CCE71937.1 JTDY01003492 KOB69475.1 GFPF01005340 MAA16486.1
Proteomes
Pfam
Interpro
IPR036084
Ser_inhib-like_sf
+ More
IPR002919 TIL_dom
IPR006212 Furin_repeat
IPR000742 EGF-like_dom
IPR004094 Antistasin-like
IPR004843 Calcineurin-like_PHP_ApaH
IPR011990 TPR-like_helical_dom_sf
IPR019734 TPR_repeat
IPR006186 Ser/Thr-sp_prot-phosphatase
IPR013026 TPR-contain_dom
IPR011236 Ser/Thr_PPase_5
IPR013235 PPP_dom
IPR029052 Metallo-depent_PP-like
IPR041753 PP5_C
IPR042185 Serpin_sf_2
IPR042178 Serpin_sf_1
IPR023796 Serpin_dom
IPR036186 Serpin_sf
IPR000361 FeS_biogenesis
IPR004855 TFIIA_asu/bsu
IPR035903 HesB-like_dom_sf
IPR016092 FeS_cluster_insertion
IPR000215 Serpin_fam
IPR023795 Serpin_CS
IPR017896 4Fe4S_Fe-S-bd
IPR002919 TIL_dom
IPR006212 Furin_repeat
IPR000742 EGF-like_dom
IPR004094 Antistasin-like
IPR004843 Calcineurin-like_PHP_ApaH
IPR011990 TPR-like_helical_dom_sf
IPR019734 TPR_repeat
IPR006186 Ser/Thr-sp_prot-phosphatase
IPR013026 TPR-contain_dom
IPR011236 Ser/Thr_PPase_5
IPR013235 PPP_dom
IPR029052 Metallo-depent_PP-like
IPR041753 PP5_C
IPR042185 Serpin_sf_2
IPR042178 Serpin_sf_1
IPR023796 Serpin_dom
IPR036186 Serpin_sf
IPR000361 FeS_biogenesis
IPR004855 TFIIA_asu/bsu
IPR035903 HesB-like_dom_sf
IPR016092 FeS_cluster_insertion
IPR000215 Serpin_fam
IPR023795 Serpin_CS
IPR017896 4Fe4S_Fe-S-bd
ProteinModelPortal
A0A2A4J071
A0A2H1VKZ4
A0A0L7LWK3
A0A2W1BA04
A0A0V0Z791
A0A2A4J6N9
+ More
A0A0V0Z7M5 A0A2A4IW13 A0A2A4IXH8 A0A0V0Z760 A0A0V1CXK4 A0A0V0Z7C1 A0A0V0S5K9 A0A0V0Z793 A0A1B0CE59 A0A0V0VIZ3 A0A0V0U5N5 A0A0V1LSL0 A0A0V0S563 E5S2I0 A0A0V1NT84 A0A0V0S515 A0A0V0U5E1 A0A0V1B9Z3 A0A0V1B9Y2 A0A2P6L3Q8 A0A1B0CYU8 A0A1J1IUR1 A0A2A4J7B9 A0A2A4J556 A0A0V0WZ71 A0A0V0WZ93 A0A0V1EPP1 A0A0V1EPN2 A0A2W1BQG2 A0A0V1J3G8 A0A0V1EPP9 H9J5D7 A0A0V1FGY8 A0A2H1VAX4 A0A0V1J3E7 A0A3G1T1R3 A0A0V1MZF6 Q8I4B9 Q9U1T5 A0A0L7L697 A0A1J1HI61 A0A0V1H1U7 H9JMY6 A0A087UJV8 A0A0K0EAF5 A0A1J1HLL5 A0A2Z6FLF1 H2L2K9 A0A0L7L220 A0A182PDJ4 A0A224YFL8
A0A0V0Z7M5 A0A2A4IW13 A0A2A4IXH8 A0A0V0Z760 A0A0V1CXK4 A0A0V0Z7C1 A0A0V0S5K9 A0A0V0Z793 A0A1B0CE59 A0A0V0VIZ3 A0A0V0U5N5 A0A0V1LSL0 A0A0V0S563 E5S2I0 A0A0V1NT84 A0A0V0S515 A0A0V0U5E1 A0A0V1B9Z3 A0A0V1B9Y2 A0A2P6L3Q8 A0A1B0CYU8 A0A1J1IUR1 A0A2A4J7B9 A0A2A4J556 A0A0V0WZ71 A0A0V0WZ93 A0A0V1EPP1 A0A0V1EPN2 A0A2W1BQG2 A0A0V1J3G8 A0A0V1EPP9 H9J5D7 A0A0V1FGY8 A0A2H1VAX4 A0A0V1J3E7 A0A3G1T1R3 A0A0V1MZF6 Q8I4B9 Q9U1T5 A0A0L7L697 A0A1J1HI61 A0A0V1H1U7 H9JMY6 A0A087UJV8 A0A0K0EAF5 A0A1J1HLL5 A0A2Z6FLF1 H2L2K9 A0A0L7L220 A0A182PDJ4 A0A224YFL8
Ontologies
GO
PANTHER
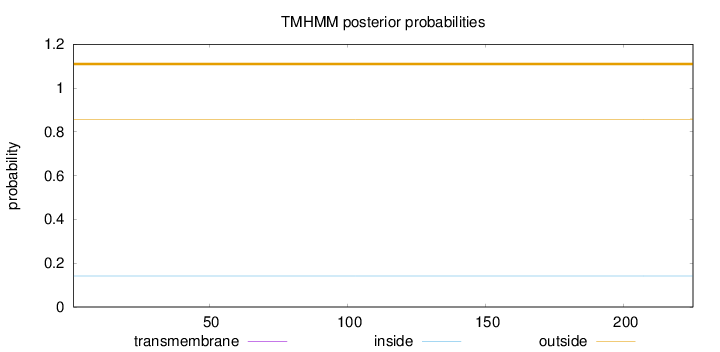
Topology
Length:
225
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00132
Exp number, first 60 AAs:
0
Total prob of N-in:
0.14204
outside
1 - 225
Population Genetic Test Statistics
Pi
160.936111
Theta
189.72746
Tajima's D
-0.612172
CLR
25.732813
CSRT
0.211139443027849
Interpretation
Uncertain
