Gene
KWMTBOMO03530
Annotation
hypothetical_protein_KGM_20753_[Danaus_plexippus]
Location in the cell
Nuclear Reliability : 1.959
Sequence
CDS
ATGCCAATTTTGTTTTCAGTCAATATACGACTCTTCGATACGGATGGCGTCCGGATCCCGGGCACGGAATGCGACTACCAGTTCAGCCGTTCCGGGAGCCGTCCGCAGCACGGCCGGCTGTACAGCCCTCGCTACCCGTCCATCTATCCCAACAACGTGCGATGTACTTATCACTTTCACGCAAGGCCAAAGGAGCGCGTGAAAGTTGTCTTCGAAGAAATTTCACTTCAAAAAGGTGACCTCAGTTGCCTCCGAAGAGCGGATATCATCAAGGTGTTTGATGGCCGGGACTCGAACGCGCCTGCGATCGCAATGCTATGCAATGAACTGACTGGTTACGAGGTGCTGTCAACGGGACCCGCACTACTGCTGCAGTTCACGGCGAATTCGGCAACACCAGGCCAGGGTTTCGCGGCCACCTTCCACTTTCAACCGCCCCCTGATTCGACGGCTGCTGATTCAGATCGTCTCCTGAAACTCAGCTTCGGTAAAGCCTTTGAATCCCTCGGGCCTGAAGTAAGCGCAACAAGTGAGTAG
Protein
MPILFSVNIRLFDTDGVRIPGTECDYQFSRSGSRPQHGRLYSPRYPSIYPNNVRCTYHFHARPKERVKVVFEEISLQKGDLSCLRRADIIKVFDGRDSNAPAIAMLCNELTGYEVLSTGPALLLQFTANSATPGQGFAATFHFQPPPDSTAADSDRLLKLSFGKAFESLGPEVSATSE
Summary
Uniprot
A0A194QQL8
A0A212EM27
A0A2W1BKR9
A0A2H1VUY5
F4WR90
A0A151JCU7
+ More
A0A158NUC7 A0A0L7LN85 A0A232FKG5 A0A154PMP2 A0A3L8DG62 A0A151IG53 A0A194QAB4 A0A194QRP3 A0A026WTQ3 A0A067RBD3 A0A0L7R497 A0A088AES6 A0A139WB16 A0A195FKA4 A0A2A3EAW5 K7JPL0 A0A0M9A963 A0A0T6B3L9 T1HQ54 A0A1D2MT37 T1JI54 A0A0A9XJS0 T1JPZ7 E9GLD3 T1JTV6 A0A0P5DCN7 A0A0P5Y957 A0A2T7P5R4 A0A034WWP8 A0A0P5LPL7 A0A0L0C0R3 A0A0P5J7C0 A0A0P5Z782 A0A0P5YSN9 A0A0P5IDP0 A0A0P5YSH4 A0A0P5YRY5 A0A0P4X3Q4 A0A0P5YSP1 A0A0N8A764 A0A0N8CFA1 A0A0P5SNR6 A0A0P5GIX3 A0A0P4YFN6 A0A0P5Y1U5 A0A0P6AKC2 A0A0B5G2C0 T1FZA9 A0A1I8N6D1 E2AB40 A0A0P6HIG0 A0A1I8PA75 A0A1I8PAC8 B4LXG7 B4GZ51 A0A267FE03 B3M2P6 A0A267GPP7 A0A0P6FV39 A0A0R3NFG4 A0A2H1CNK7 B4JRF3 B4KAX9 A0A0M4EDT9 A0A183A5J1 B3P1C0 B4HGZ4 A0A2B4RH88 B4NBE2
A0A158NUC7 A0A0L7LN85 A0A232FKG5 A0A154PMP2 A0A3L8DG62 A0A151IG53 A0A194QAB4 A0A194QRP3 A0A026WTQ3 A0A067RBD3 A0A0L7R497 A0A088AES6 A0A139WB16 A0A195FKA4 A0A2A3EAW5 K7JPL0 A0A0M9A963 A0A0T6B3L9 T1HQ54 A0A1D2MT37 T1JI54 A0A0A9XJS0 T1JPZ7 E9GLD3 T1JTV6 A0A0P5DCN7 A0A0P5Y957 A0A2T7P5R4 A0A034WWP8 A0A0P5LPL7 A0A0L0C0R3 A0A0P5J7C0 A0A0P5Z782 A0A0P5YSN9 A0A0P5IDP0 A0A0P5YSH4 A0A0P5YRY5 A0A0P4X3Q4 A0A0P5YSP1 A0A0N8A764 A0A0N8CFA1 A0A0P5SNR6 A0A0P5GIX3 A0A0P4YFN6 A0A0P5Y1U5 A0A0P6AKC2 A0A0B5G2C0 T1FZA9 A0A1I8N6D1 E2AB40 A0A0P6HIG0 A0A1I8PA75 A0A1I8PAC8 B4LXG7 B4GZ51 A0A267FE03 B3M2P6 A0A267GPP7 A0A0P6FV39 A0A0R3NFG4 A0A2H1CNK7 B4JRF3 B4KAX9 A0A0M4EDT9 A0A183A5J1 B3P1C0 B4HGZ4 A0A2B4RH88 B4NBE2
Pubmed
EMBL
KQ461181
KPJ07644.1
AGBW02013948
OWR42527.1
KZ149987
PZC75672.1
+ More
ODYU01004586 SOQ44618.1 GL888284 EGI63307.1 KQ979048 KYN23213.1 ADTU01026344 ADTU01026345 ADTU01026346 ADTU01026347 ADTU01026348 ADTU01026349 ADTU01026350 ADTU01026351 ADTU01026352 ADTU01026353 JTDY01000474 KOB77013.1 NNAY01000074 OXU31246.1 KQ434948 KZC12480.1 QOIP01000008 RLU19163.1 KQ977739 KYN00204.1 KQ459249 KPJ02417.1 KPJ07650.1 KK107109 EZA59338.1 KK852752 KDR17108.1 KQ414657 KOC65710.1 KQ971374 KYB25138.1 KQ981512 KYN40833.1 KZ288301 PBC28830.1 AAZX01000463 AAZX01012136 AAZX01012966 KQ435710 KOX79767.1 LJIG01015992 KRT81938.1 ACPB03022776 LJIJ01000588 ODM96058.1 JH431584 GBHO01022637 JAG20967.1 CAEY01000435 CAEY01000436 GL732551 EFX79492.1 CAEY01000486 GDIP01163274 JAJ60128.1 GDIP01074678 JAM29037.1 PZQS01000006 PVD28760.1 GAKP01000200 JAC58752.1 GDIQ01167745 JAK83980.1 JRES01001160 KNC25049.1 GDIQ01205503 JAK46222.1 GDIP01048133 JAM55582.1 GDIP01053938 JAM49777.1 GDIQ01222388 JAK29337.1 GDIP01053939 JAM49776.1 GDIP01053937 JAM49778.1 GDIP01249765 JAI73636.1 GDIP01053936 JAM49779.1 GDIP01175825 JAJ47577.1 GDIP01137439 JAL66275.1 GDIP01137438 JAL66276.1 GDIQ01239881 JAK11844.1 GDIP01228271 JAI95130.1 GDIP01065774 JAM37941.1 GDIP01028198 JAM75517.1 KP067952 AJF11714.1 AMQM01001384 KB097495 ESN96367.1 GL438234 EFN69316.1 GDIQ01018881 JAN75856.1 CH940650 EDW67845.2 CH479198 EDW28069.1 NIVC01001179 PAA71232.1 CH902617 EDV42367.2 NIVC01000240 PAA87262.1 GDIQ01053311 JAN41426.1 CM000070 KRS99808.1 KZ428827 PIS89044.1 CH916373 EDV94343.1 CH933806 EDW14657.2 CP012526 ALC46041.1 UZAN01039481 VDP65808.1 CH954181 EDV49379.1 CH480815 EDW42465.1 LSMT01000505 PFX16981.1 CH964232 EDW81106.2
ODYU01004586 SOQ44618.1 GL888284 EGI63307.1 KQ979048 KYN23213.1 ADTU01026344 ADTU01026345 ADTU01026346 ADTU01026347 ADTU01026348 ADTU01026349 ADTU01026350 ADTU01026351 ADTU01026352 ADTU01026353 JTDY01000474 KOB77013.1 NNAY01000074 OXU31246.1 KQ434948 KZC12480.1 QOIP01000008 RLU19163.1 KQ977739 KYN00204.1 KQ459249 KPJ02417.1 KPJ07650.1 KK107109 EZA59338.1 KK852752 KDR17108.1 KQ414657 KOC65710.1 KQ971374 KYB25138.1 KQ981512 KYN40833.1 KZ288301 PBC28830.1 AAZX01000463 AAZX01012136 AAZX01012966 KQ435710 KOX79767.1 LJIG01015992 KRT81938.1 ACPB03022776 LJIJ01000588 ODM96058.1 JH431584 GBHO01022637 JAG20967.1 CAEY01000435 CAEY01000436 GL732551 EFX79492.1 CAEY01000486 GDIP01163274 JAJ60128.1 GDIP01074678 JAM29037.1 PZQS01000006 PVD28760.1 GAKP01000200 JAC58752.1 GDIQ01167745 JAK83980.1 JRES01001160 KNC25049.1 GDIQ01205503 JAK46222.1 GDIP01048133 JAM55582.1 GDIP01053938 JAM49777.1 GDIQ01222388 JAK29337.1 GDIP01053939 JAM49776.1 GDIP01053937 JAM49778.1 GDIP01249765 JAI73636.1 GDIP01053936 JAM49779.1 GDIP01175825 JAJ47577.1 GDIP01137439 JAL66275.1 GDIP01137438 JAL66276.1 GDIQ01239881 JAK11844.1 GDIP01228271 JAI95130.1 GDIP01065774 JAM37941.1 GDIP01028198 JAM75517.1 KP067952 AJF11714.1 AMQM01001384 KB097495 ESN96367.1 GL438234 EFN69316.1 GDIQ01018881 JAN75856.1 CH940650 EDW67845.2 CH479198 EDW28069.1 NIVC01001179 PAA71232.1 CH902617 EDV42367.2 NIVC01000240 PAA87262.1 GDIQ01053311 JAN41426.1 CM000070 KRS99808.1 KZ428827 PIS89044.1 CH916373 EDV94343.1 CH933806 EDW14657.2 CP012526 ALC46041.1 UZAN01039481 VDP65808.1 CH954181 EDV49379.1 CH480815 EDW42465.1 LSMT01000505 PFX16981.1 CH964232 EDW81106.2
Proteomes
UP000053240
UP000007151
UP000007755
UP000078492
UP000005205
UP000037510
+ More
UP000215335 UP000076502 UP000279307 UP000078542 UP000053268 UP000053097 UP000027135 UP000053825 UP000005203 UP000007266 UP000078541 UP000242457 UP000002358 UP000053105 UP000015103 UP000094527 UP000015104 UP000000305 UP000245119 UP000037069 UP000015101 UP000095301 UP000000311 UP000095300 UP000008792 UP000008744 UP000215902 UP000007801 UP000001819 UP000001070 UP000009192 UP000092553 UP000050740 UP000008711 UP000001292 UP000225706 UP000007798
UP000215335 UP000076502 UP000279307 UP000078542 UP000053268 UP000053097 UP000027135 UP000053825 UP000005203 UP000007266 UP000078541 UP000242457 UP000002358 UP000053105 UP000015103 UP000094527 UP000015104 UP000000305 UP000245119 UP000037069 UP000015101 UP000095301 UP000000311 UP000095300 UP000008792 UP000008744 UP000215902 UP000007801 UP000001819 UP000001070 UP000009192 UP000092553 UP000050740 UP000008711 UP000001292 UP000225706 UP000007798
Interpro
Gene 3D
ProteinModelPortal
A0A194QQL8
A0A212EM27
A0A2W1BKR9
A0A2H1VUY5
F4WR90
A0A151JCU7
+ More
A0A158NUC7 A0A0L7LN85 A0A232FKG5 A0A154PMP2 A0A3L8DG62 A0A151IG53 A0A194QAB4 A0A194QRP3 A0A026WTQ3 A0A067RBD3 A0A0L7R497 A0A088AES6 A0A139WB16 A0A195FKA4 A0A2A3EAW5 K7JPL0 A0A0M9A963 A0A0T6B3L9 T1HQ54 A0A1D2MT37 T1JI54 A0A0A9XJS0 T1JPZ7 E9GLD3 T1JTV6 A0A0P5DCN7 A0A0P5Y957 A0A2T7P5R4 A0A034WWP8 A0A0P5LPL7 A0A0L0C0R3 A0A0P5J7C0 A0A0P5Z782 A0A0P5YSN9 A0A0P5IDP0 A0A0P5YSH4 A0A0P5YRY5 A0A0P4X3Q4 A0A0P5YSP1 A0A0N8A764 A0A0N8CFA1 A0A0P5SNR6 A0A0P5GIX3 A0A0P4YFN6 A0A0P5Y1U5 A0A0P6AKC2 A0A0B5G2C0 T1FZA9 A0A1I8N6D1 E2AB40 A0A0P6HIG0 A0A1I8PA75 A0A1I8PAC8 B4LXG7 B4GZ51 A0A267FE03 B3M2P6 A0A267GPP7 A0A0P6FV39 A0A0R3NFG4 A0A2H1CNK7 B4JRF3 B4KAX9 A0A0M4EDT9 A0A183A5J1 B3P1C0 B4HGZ4 A0A2B4RH88 B4NBE2
A0A158NUC7 A0A0L7LN85 A0A232FKG5 A0A154PMP2 A0A3L8DG62 A0A151IG53 A0A194QAB4 A0A194QRP3 A0A026WTQ3 A0A067RBD3 A0A0L7R497 A0A088AES6 A0A139WB16 A0A195FKA4 A0A2A3EAW5 K7JPL0 A0A0M9A963 A0A0T6B3L9 T1HQ54 A0A1D2MT37 T1JI54 A0A0A9XJS0 T1JPZ7 E9GLD3 T1JTV6 A0A0P5DCN7 A0A0P5Y957 A0A2T7P5R4 A0A034WWP8 A0A0P5LPL7 A0A0L0C0R3 A0A0P5J7C0 A0A0P5Z782 A0A0P5YSN9 A0A0P5IDP0 A0A0P5YSH4 A0A0P5YRY5 A0A0P4X3Q4 A0A0P5YSP1 A0A0N8A764 A0A0N8CFA1 A0A0P5SNR6 A0A0P5GIX3 A0A0P4YFN6 A0A0P5Y1U5 A0A0P6AKC2 A0A0B5G2C0 T1FZA9 A0A1I8N6D1 E2AB40 A0A0P6HIG0 A0A1I8PA75 A0A1I8PAC8 B4LXG7 B4GZ51 A0A267FE03 B3M2P6 A0A267GPP7 A0A0P6FV39 A0A0R3NFG4 A0A2H1CNK7 B4JRF3 B4KAX9 A0A0M4EDT9 A0A183A5J1 B3P1C0 B4HGZ4 A0A2B4RH88 B4NBE2
PDB
2QQO
E-value=2.31291e-08,
Score=135
Ontologies
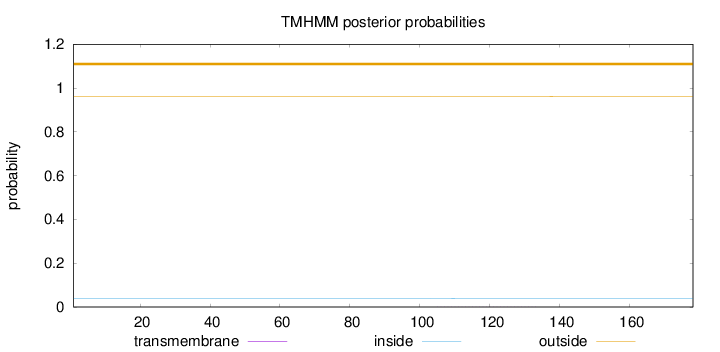
Topology
Length:
178
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00874
Exp number, first 60 AAs:
0
Total prob of N-in:
0.03952
outside
1 - 178
Population Genetic Test Statistics
Pi
191.690855
Theta
171.696982
Tajima's D
0.100201
CLR
0.600527
CSRT
0.390730463476826
Interpretation
Uncertain
