Gene
KWMTBOMO03527
Annotation
reverse_transcriptase_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.333
Sequence
CDS
ATGCGTGCCCTACAACGCGACGTAAAGTCTCGCATCGCCGAAGTCCGAGATGCCAGATGGTCTGATTTCTTAGAAGGACTCGCACCCTCCCATAGGTCTTACTACCGCTTAGCTCGTACTCTCAAATCGGATACGGTAGTAACTATGCCCCCCCTCGTAGGCCCCTCAGGCCGACTCGCGGCGTTCGATGATGACGAAAAAGCAGAGCTGCTGGCCGATACATTGCAAACCCAGTGCACGCCCAGCACTCAATCCGTGGACCCTGTTCATGTAGAATTAATAGACAGTGAGGTAGAACGCAGAGCCTCCTTGCCACCATCGGATGCGTTACCACCCGTCACCCCGATAGAAGTTAAAGACTTGATCAAAGACCTACGTCCTCGCAAGGCTCCCGGTTCCGACGGTATATCTAACCGCGTTATTAAACTTCTACCCGTCCAACTCATCGTGATGTTGGCATCTATTTTCAATGCCGCTATGGCGAACTGTATCTTTCCCGCGGTAAACGGAGCGTACGTGGTGAAGAGAGACACACCATCGACCCCCCCGAATATATTTGAATCATTTCAAACAAACATGGACATGTTCAGGAAGTGCTTGGATTTAACGCTGGCCAAATCGGTGGACGATGTTAACGTGCATCAACTGAAACCTGTCTTTAATGTTATCGGTGATCAGATTAATAGATTTTCTAAAGCGTTCGAAGATTTGACCGCGAAAAGTACTACAACACAGGAATCGATTTGGGCGTAG
Protein
MRALQRDVKSRIAEVRDARWSDFLEGLAPSHRSYYRLARTLKSDTVVTMPPLVGPSGRLAAFDDDEKAELLADTLQTQCTPSTQSVDPVHVELIDSEVERRASLPPSDALPPVTPIEVKDLIKDLRPRKAPGSDGISNRVIKLLPVQLIVMLASIFNAAMANCIFPAVNGAYVVKRDTPSTPPNIFESFQTNMDMFRKCLDLTLAKSVDDVNVHQLKPVFNVIGDQINRFSKAFEDLTAKSTTTQESIWA
Summary
Uniprot
Q93137
Q9XXW0
Q9BPQ0
A0A0N1PFV1
A0A1Y1NA23
A0A2S2Q352
+ More
A0A1B6J5Z9 A0A2S2PEZ9 A0A2S2Q5V3 A0A2H8U0H5 X1WKS2 K7IN67 A0A087UB64 A0A2S2QEE0 A0A2S2NPT6 A0A2S2R7N0 J9KLB1 A0A2L2YNG9 A0A2L2YN79 A0A3N5Z7I3 A0A2S2N6C8 A0A2S2N629 A0A2S2PA10 A0A224XI99 T1ID59 A0A2S2PBB4 A0A023F700 A0A2J7R7D0 Q07995 J9K7I4 A0A2S2RAN7 A0A224X6U5 X1WVF0
A0A1B6J5Z9 A0A2S2PEZ9 A0A2S2Q5V3 A0A2H8U0H5 X1WKS2 K7IN67 A0A087UB64 A0A2S2QEE0 A0A2S2NPT6 A0A2S2R7N0 J9KLB1 A0A2L2YNG9 A0A2L2YN79 A0A3N5Z7I3 A0A2S2N6C8 A0A2S2N629 A0A2S2PA10 A0A224XI99 T1ID59 A0A2S2PBB4 A0A023F700 A0A2J7R7D0 Q07995 J9K7I4 A0A2S2RAN7 A0A224X6U5 X1WVF0
EMBL
U07847
AAA17752.1
AB018558
BAA76304.1
AB055391
BAB21761.1
+ More
KQ459265 KPJ02064.1 GEZM01008457 JAV94772.1 GGMS01002955 MBY72158.1 GECU01013097 JAS94609.1 GGMR01015412 MBY28031.1 GGMS01003914 MBY73117.1 GFXV01007093 MBW18898.1 ABLF02005245 ABLF02013477 ABLF02047385 ABLF02066435 KK119069 KFM74603.1 GGMS01006359 MBY75562.1 GGMR01006581 MBY19200.1 GGMS01016818 MBY86021.1 ABLF02040174 ABLF02040183 ABLF02055266 IAAA01035748 LAA09609.1 IAAA01035749 LAA09611.1 RPOV01000134 RPJ78669.1 GGMR01000086 MBY12705.1 GGMR01000005 MBY12624.1 GGMR01013409 MBY26028.1 GFTR01008256 JAW08170.1 ACPB03027378 GGMR01014053 MBY26672.1 GBBI01001699 JAC17013.1 NEVH01006736 PNF36716.1 S59870 AAB26437.2 ABLF02017543 ABLF02017549 ABLF02017551 ABLF02021996 GGMS01017914 MBY87117.1 GFTR01008251 JAW08175.1 ABLF02003375 ABLF02032497
KQ459265 KPJ02064.1 GEZM01008457 JAV94772.1 GGMS01002955 MBY72158.1 GECU01013097 JAS94609.1 GGMR01015412 MBY28031.1 GGMS01003914 MBY73117.1 GFXV01007093 MBW18898.1 ABLF02005245 ABLF02013477 ABLF02047385 ABLF02066435 KK119069 KFM74603.1 GGMS01006359 MBY75562.1 GGMR01006581 MBY19200.1 GGMS01016818 MBY86021.1 ABLF02040174 ABLF02040183 ABLF02055266 IAAA01035748 LAA09609.1 IAAA01035749 LAA09611.1 RPOV01000134 RPJ78669.1 GGMR01000086 MBY12705.1 GGMR01000005 MBY12624.1 GGMR01013409 MBY26028.1 GFTR01008256 JAW08170.1 ACPB03027378 GGMR01014053 MBY26672.1 GBBI01001699 JAC17013.1 NEVH01006736 PNF36716.1 S59870 AAB26437.2 ABLF02017543 ABLF02017549 ABLF02017551 ABLF02021996 GGMS01017914 MBY87117.1 GFTR01008251 JAW08175.1 ABLF02003375 ABLF02032497
Proteomes
Pfam
Interpro
IPR036691
Endo/exonu/phosph_ase_sf
+ More
IPR005135 Endo/exonuclease/phosphatase
IPR000477 RT_dom
IPR006579 Pre_C2HC_dom
IPR041588 Integrase_H2C2
IPR036397 RNaseH_sf
IPR005312 DUF1759
IPR040676 DUF5641
IPR008737 Peptidase_asp_put
IPR008042 Retrotrans_Pao
IPR012337 RNaseH-like_sf
IPR001584 Integrase_cat-core
IPR005135 Endo/exonuclease/phosphatase
IPR000477 RT_dom
IPR006579 Pre_C2HC_dom
IPR041588 Integrase_H2C2
IPR036397 RNaseH_sf
IPR005312 DUF1759
IPR040676 DUF5641
IPR008737 Peptidase_asp_put
IPR008042 Retrotrans_Pao
IPR012337 RNaseH-like_sf
IPR001584 Integrase_cat-core
Gene 3D
ProteinModelPortal
Q93137
Q9XXW0
Q9BPQ0
A0A0N1PFV1
A0A1Y1NA23
A0A2S2Q352
+ More
A0A1B6J5Z9 A0A2S2PEZ9 A0A2S2Q5V3 A0A2H8U0H5 X1WKS2 K7IN67 A0A087UB64 A0A2S2QEE0 A0A2S2NPT6 A0A2S2R7N0 J9KLB1 A0A2L2YNG9 A0A2L2YN79 A0A3N5Z7I3 A0A2S2N6C8 A0A2S2N629 A0A2S2PA10 A0A224XI99 T1ID59 A0A2S2PBB4 A0A023F700 A0A2J7R7D0 Q07995 J9K7I4 A0A2S2RAN7 A0A224X6U5 X1WVF0
A0A1B6J5Z9 A0A2S2PEZ9 A0A2S2Q5V3 A0A2H8U0H5 X1WKS2 K7IN67 A0A087UB64 A0A2S2QEE0 A0A2S2NPT6 A0A2S2R7N0 J9KLB1 A0A2L2YNG9 A0A2L2YN79 A0A3N5Z7I3 A0A2S2N6C8 A0A2S2N629 A0A2S2PA10 A0A224XI99 T1ID59 A0A2S2PBB4 A0A023F700 A0A2J7R7D0 Q07995 J9K7I4 A0A2S2RAN7 A0A224X6U5 X1WVF0
Ontologies
Topology
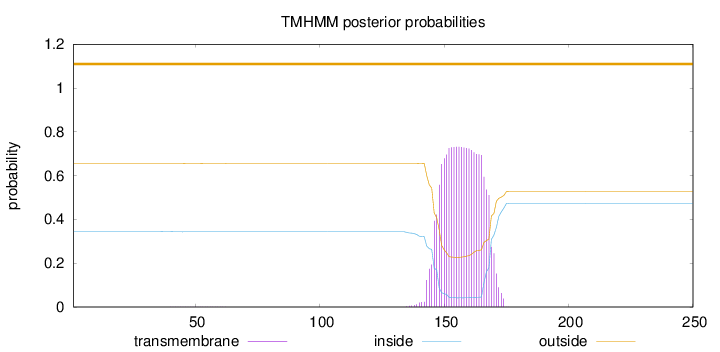
Length:
250
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
16.58744
Exp number, first 60 AAs:
0.0192
Total prob of N-in:
0.34427
outside
1 - 250
Population Genetic Test Statistics
Pi
2.928025
Theta
35.057778
Tajima's D
-1.053662
CLR
0.348392
CSRT
0.127193640317984
Interpretation
Uncertain
