Pre Gene Modal
BGIBMGA006246
Annotation
PREDICTED:_NADP-dependent_malic_enzyme_[Bombyx_mori]
Full name
Malic enzyme
Location in the cell
Mitochondrial Reliability : 1.339
Sequence
CDS
ATGTTTTCCAGTGTGTCCAGATCGTTATTATGTTGCACAAACGCAATAGGTCCAAACAGGGCCCTGGCGTACATCCTAACAAAGAACTCGATCCTCGAACCTTCCTCGATACAATGTCGGGATTACCACGAAGTAACCGGCAATATCATTTGCCCGAATGTTGTCCGAGGCATCGACCACCTTAGAGATCCTCGCCTTAATAAAGGTCTCGCGTTTACTTTAGAAGAACGTCAGGCTCTCGGCATTCACGGTCTTTTGGCGGCCAAATTCAAAAGTCAAGAAGAACAGCTGGACATTTGCAGGATATCAGTGGACAGATACGAGGACAATCTGAACAAATATCTCTACCTCGTCGAGTTACAGGATCGAAACGAGAAACTGTTCTTTAGACTTCTAGCCGAGAACGTCGAGAAGTATTTACCGATAGTGTACACTCCGACCGTTGGTCTGGCCTGTCAGAGGTTCGGCCTCATATACCGAAGGCCGAGAGGTCTCTTCGTGACTATCAACGACAAAGGCCACGTGTTTAACATCATAAAAAATTGGCCAGAACCTGATGTCCGAGCGATCGTTTTCACCGATGGCGAGAGAATATTGGGCTTGGGTGATCTCGGAGCGTACGGTATGGGGATTCCAGTGGGAAAGCTGTCGCTTTACACAGCTCTGGCTGGGATTAAGCCACACCAATGCTTACCGATTACGCTGGACGTCGGAACGGACAATCAGGTAAAACTTGAAACCATGGCCTAA
Protein
MFSSVSRSLLCCTNAIGPNRALAYILTKNSILEPSSIQCRDYHEVTGNIICPNVVRGIDHLRDPRLNKGLAFTLEERQALGIHGLLAAKFKSQEEQLDICRISVDRYEDNLNKYLYLVELQDRNEKLFFRLLAENVEKYLPIVYTPTVGLACQRFGLIYRRPRGLFVTINDKGHVFNIIKNWPEPDVRAIVFTDGERILGLGDLGAYGMGIPVGKLSLYTALAGIKPHQCLPITLDVGTDNQVKLETMA
Summary
Similarity
Belongs to the malic enzymes family.
Belongs to the peptidase C19 family.
Belongs to the peptidase C19 family.
Uniprot
H9J9Q3
A0A2W1C0I7
A0A194QBR2
A0A2A4JIT3
A0A2H1V9X6
A0A194QVQ3
+ More
A0A212F922 A0A182SMD5 A0A2J7Q031 F5HL44 A0A182XJ28 A0A182V0I8 A0A182I7T1 A0A1B6LG77 A0A182P1Q3 A0A182LYI4 T1E996 A0A067QF27 A0A182TL51 A0A182YGM6 A0A182W1I7 A0A182IQ03 A0A182K1D7 T1PDX1 A0A182RYM9 T1PDU2 A0A182R0A2 A0A182N5S8 A0A2M4ABQ9 A0A2M4ABS9 W5JPA9 A0A182FPU6 A0A1L8E0R1 A0A0K8TU30 A0A2H8TEF4 A0A1S4EXR8 A0A1B6H4G2 Q17M99 A0A2M4BHA3 A0A2M4BHC5 A0A182H086 A0A182GYQ8 A0A2M3Z470 A0A1I8NSM9 A0A2M3Z419 U5EUA8 D6WC30 B0WQC9 A0A336MLA2 A0A1B6DG24 A0A1B6C744 A0A023EXF3 A0A1Q3FI86 A0A1B0DBG6 A0A2P8XVF2 V5GLS7 A0A1B6K6A7 A0A0A9XJW4 J9K007 A0A2H8TYS0 A0A2S2NBL0 A0A0L0BYF3 B4PRU5 T1I5N5 B4IC50 A0A0R1E5P7 A0A2S2Q9I8 A0A023F4G0 A0A0V0GBL2 A0A1W4UIU7 H9J9Q4 A0A2J7Q043 B4KAX8 A0A3B0KAI4 A0A0Q9WYS2 A0A1W4UH31 B4NBE9 A0A0P4VX09 A0A0M5J924 A0A1J1I538 A0A3B0K3M4 A0A0Q9WT09 A0A3B0JE23 B4G2G6 B4JU20 B4LXG6 A0A0P4W311 A0A0Q9WG72 Q9U1J0 A0A1B0B2N3 B3P850 Q9VB69 A0A0P4W9N3 E1JIZ4 E1JIZ5 W8BCJ7 A0A1A9YSV7 A0A1B0FEH5 Q29AC6 A0A1A9ZZ14
A0A212F922 A0A182SMD5 A0A2J7Q031 F5HL44 A0A182XJ28 A0A182V0I8 A0A182I7T1 A0A1B6LG77 A0A182P1Q3 A0A182LYI4 T1E996 A0A067QF27 A0A182TL51 A0A182YGM6 A0A182W1I7 A0A182IQ03 A0A182K1D7 T1PDX1 A0A182RYM9 T1PDU2 A0A182R0A2 A0A182N5S8 A0A2M4ABQ9 A0A2M4ABS9 W5JPA9 A0A182FPU6 A0A1L8E0R1 A0A0K8TU30 A0A2H8TEF4 A0A1S4EXR8 A0A1B6H4G2 Q17M99 A0A2M4BHA3 A0A2M4BHC5 A0A182H086 A0A182GYQ8 A0A2M3Z470 A0A1I8NSM9 A0A2M3Z419 U5EUA8 D6WC30 B0WQC9 A0A336MLA2 A0A1B6DG24 A0A1B6C744 A0A023EXF3 A0A1Q3FI86 A0A1B0DBG6 A0A2P8XVF2 V5GLS7 A0A1B6K6A7 A0A0A9XJW4 J9K007 A0A2H8TYS0 A0A2S2NBL0 A0A0L0BYF3 B4PRU5 T1I5N5 B4IC50 A0A0R1E5P7 A0A2S2Q9I8 A0A023F4G0 A0A0V0GBL2 A0A1W4UIU7 H9J9Q4 A0A2J7Q043 B4KAX8 A0A3B0KAI4 A0A0Q9WYS2 A0A1W4UH31 B4NBE9 A0A0P4VX09 A0A0M5J924 A0A1J1I538 A0A3B0K3M4 A0A0Q9WT09 A0A3B0JE23 B4G2G6 B4JU20 B4LXG6 A0A0P4W311 A0A0Q9WG72 Q9U1J0 A0A1B0B2N3 B3P850 Q9VB69 A0A0P4W9N3 E1JIZ4 E1JIZ5 W8BCJ7 A0A1A9YSV7 A0A1B0FEH5 Q29AC6 A0A1A9ZZ14
Pubmed
19121390
28756777
26354079
22118469
12364791
14747013
+ More
17210077 24845553 25244985 25315136 20920257 23761445 26369729 17510324 26483478 18362917 19820115 24945155 29403074 25401762 26823975 26108605 17994087 17550304 25474469 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24495485 15632085
17210077 24845553 25244985 25315136 20920257 23761445 26369729 17510324 26483478 18362917 19820115 24945155 29403074 25401762 26823975 26108605 17994087 17550304 25474469 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24495485 15632085
EMBL
BABH01034014
BABH01034015
BABH01034016
BABH01034017
KZ149899
PZC78630.1
+ More
KQ459249 KPJ02430.1 NWSH01001337 PCG71618.1 ODYU01001426 SOQ37596.1 KQ461181 KPJ07636.1 AGBW02009653 OWR50228.1 NEVH01020327 PNF21947.1 AAAB01008880 EGK97005.1 APCN01002552 GEBQ01017403 JAT22574.1 AXCM01000270 GAMD01002043 JAA99547.1 KK853743 KDQ87031.1 KA646941 AFP61570.1 KA646942 AFP61571.1 AXCN02001157 GGFK01004902 MBW38223.1 GGFK01004916 MBW38237.1 ADMH02000944 ETN64604.1 GFDF01001848 JAV12236.1 GDAI01000138 JAI17465.1 GFXV01000666 MBW12471.1 GECZ01000197 JAS69572.1 CH477207 EAT47850.1 GGFJ01003263 MBW52404.1 GGFJ01003314 MBW52455.1 JXUM01100888 JXUM01100889 KQ564606 KXJ72035.1 JXUM01021496 JXUM01021497 JXUM01021498 KQ560602 KXJ81680.1 GGFM01002542 MBW23293.1 GGFM01002520 MBW23271.1 GANO01001567 JAB58304.1 KQ971310 EEZ99125.1 DS232039 EDS32800.1 UFQS01001564 UFQT01001564 SSX11469.1 SSX31036.1 GEDC01012664 JAS24634.1 GEDC01028213 JAS09085.1 GAPW01000494 JAC13104.1 GFDL01007813 JAV27232.1 AJVK01029972 PYGN01001288 PSN35985.1 GALX01003447 JAB65019.1 GECU01000718 JAT06989.1 GBHO01023663 GBRD01005565 GDHC01002619 JAG19941.1 JAG60256.1 JAQ16010.1 ABLF02026860 ABLF02026861 GFXV01007658 MBW19463.1 GGMR01001713 MBY14332.1 JRES01001160 KNC25050.1 CM000160 EDW98538.1 ACPB03014452 CH480828 EDW45208.1 KRK04335.1 GGMS01005184 MBY74387.1 GBBI01002297 JAC16415.1 GECL01000725 JAP05399.1 BABH01034018 PNF21946.1 CH933806 EDW14656.1 OUUW01000005 SPP80578.1 KRG01135.1 CH964232 EDW81113.1 GDRN01107055 JAI57478.1 CP012526 ALC46036.1 CVRI01000041 CRK95392.1 SPP80579.1 KRF99390.1 SPP80577.1 CH479179 EDW24011.1 CH916374 EDV91599.1 CH940650 EDW67844.1 GDRN01107058 JAI57477.1 KRF83517.1 AJ251546 CAB64263.1 JXJN01007685 CH954182 EDV53454.1 AE014297 AY051465 AAF56674.1 AAK92889.1 GDRN01107060 JAI57476.1 ACZ95044.1 ACZ95043.1 GAMC01019021 JAB87534.1 CCAG010001986 CM000070 EAL27424.3
KQ459249 KPJ02430.1 NWSH01001337 PCG71618.1 ODYU01001426 SOQ37596.1 KQ461181 KPJ07636.1 AGBW02009653 OWR50228.1 NEVH01020327 PNF21947.1 AAAB01008880 EGK97005.1 APCN01002552 GEBQ01017403 JAT22574.1 AXCM01000270 GAMD01002043 JAA99547.1 KK853743 KDQ87031.1 KA646941 AFP61570.1 KA646942 AFP61571.1 AXCN02001157 GGFK01004902 MBW38223.1 GGFK01004916 MBW38237.1 ADMH02000944 ETN64604.1 GFDF01001848 JAV12236.1 GDAI01000138 JAI17465.1 GFXV01000666 MBW12471.1 GECZ01000197 JAS69572.1 CH477207 EAT47850.1 GGFJ01003263 MBW52404.1 GGFJ01003314 MBW52455.1 JXUM01100888 JXUM01100889 KQ564606 KXJ72035.1 JXUM01021496 JXUM01021497 JXUM01021498 KQ560602 KXJ81680.1 GGFM01002542 MBW23293.1 GGFM01002520 MBW23271.1 GANO01001567 JAB58304.1 KQ971310 EEZ99125.1 DS232039 EDS32800.1 UFQS01001564 UFQT01001564 SSX11469.1 SSX31036.1 GEDC01012664 JAS24634.1 GEDC01028213 JAS09085.1 GAPW01000494 JAC13104.1 GFDL01007813 JAV27232.1 AJVK01029972 PYGN01001288 PSN35985.1 GALX01003447 JAB65019.1 GECU01000718 JAT06989.1 GBHO01023663 GBRD01005565 GDHC01002619 JAG19941.1 JAG60256.1 JAQ16010.1 ABLF02026860 ABLF02026861 GFXV01007658 MBW19463.1 GGMR01001713 MBY14332.1 JRES01001160 KNC25050.1 CM000160 EDW98538.1 ACPB03014452 CH480828 EDW45208.1 KRK04335.1 GGMS01005184 MBY74387.1 GBBI01002297 JAC16415.1 GECL01000725 JAP05399.1 BABH01034018 PNF21946.1 CH933806 EDW14656.1 OUUW01000005 SPP80578.1 KRG01135.1 CH964232 EDW81113.1 GDRN01107055 JAI57478.1 CP012526 ALC46036.1 CVRI01000041 CRK95392.1 SPP80579.1 KRF99390.1 SPP80577.1 CH479179 EDW24011.1 CH916374 EDV91599.1 CH940650 EDW67844.1 GDRN01107058 JAI57477.1 KRF83517.1 AJ251546 CAB64263.1 JXJN01007685 CH954182 EDV53454.1 AE014297 AY051465 AAF56674.1 AAK92889.1 GDRN01107060 JAI57476.1 ACZ95044.1 ACZ95043.1 GAMC01019021 JAB87534.1 CCAG010001986 CM000070 EAL27424.3
Proteomes
UP000005204
UP000053268
UP000218220
UP000053240
UP000007151
UP000075901
+ More
UP000235965 UP000007062 UP000076407 UP000075903 UP000075840 UP000075885 UP000075883 UP000027135 UP000075902 UP000076408 UP000075920 UP000075880 UP000075881 UP000095301 UP000075900 UP000075886 UP000075884 UP000000673 UP000069272 UP000008820 UP000069940 UP000249989 UP000095300 UP000007266 UP000002320 UP000092462 UP000245037 UP000007819 UP000037069 UP000002282 UP000015103 UP000001292 UP000192221 UP000009192 UP000268350 UP000007798 UP000092553 UP000183832 UP000008744 UP000001070 UP000008792 UP000092460 UP000008711 UP000000803 UP000092443 UP000092444 UP000001819 UP000092445
UP000235965 UP000007062 UP000076407 UP000075903 UP000075840 UP000075885 UP000075883 UP000027135 UP000075902 UP000076408 UP000075920 UP000075880 UP000075881 UP000095301 UP000075900 UP000075886 UP000075884 UP000000673 UP000069272 UP000008820 UP000069940 UP000249989 UP000095300 UP000007266 UP000002320 UP000092462 UP000245037 UP000007819 UP000037069 UP000002282 UP000015103 UP000001292 UP000192221 UP000009192 UP000268350 UP000007798 UP000092553 UP000183832 UP000008744 UP000001070 UP000008792 UP000092460 UP000008711 UP000000803 UP000092443 UP000092444 UP000001819 UP000092445
Pfam
Interpro
IPR037062
Malic_N_dom_sf
+ More
IPR001891 Malic_OxRdtase
IPR012302 Malic_NAD-bd
IPR015884 Malic_enzyme_CS
IPR012301 Malic_N_dom
IPR036291 NAD(P)-bd_dom_sf
IPR035914 Sperma_CUB_dom_sf
IPR000859 CUB_dom
IPR016024 ARM-type_fold
IPR015341 Glyco_hydro_38_cen
IPR011013 Gal_mutarotase_sf_dom
IPR000602 Glyco_hydro_38_N
IPR011330 Glyco_hydro/deAcase_b/a-brl
IPR028889 USP_dom
IPR037094 Glyco_hydro_38_cen_sf
IPR027291 Glyco_hydro_38_N_sf
IPR028995 Glyco_hydro_57/38_cen_sf
IPR038765 Papain-like_cys_pep_sf
IPR018200 USP_CS
IPR011989 ARM-like
IPR013780 Glyco_hydro_b
IPR011682 Glyco_hydro_38_C
IPR001394 Peptidase_C19_UCH
IPR001891 Malic_OxRdtase
IPR012302 Malic_NAD-bd
IPR015884 Malic_enzyme_CS
IPR012301 Malic_N_dom
IPR036291 NAD(P)-bd_dom_sf
IPR035914 Sperma_CUB_dom_sf
IPR000859 CUB_dom
IPR016024 ARM-type_fold
IPR015341 Glyco_hydro_38_cen
IPR011013 Gal_mutarotase_sf_dom
IPR000602 Glyco_hydro_38_N
IPR011330 Glyco_hydro/deAcase_b/a-brl
IPR028889 USP_dom
IPR037094 Glyco_hydro_38_cen_sf
IPR027291 Glyco_hydro_38_N_sf
IPR028995 Glyco_hydro_57/38_cen_sf
IPR038765 Papain-like_cys_pep_sf
IPR018200 USP_CS
IPR011989 ARM-like
IPR013780 Glyco_hydro_b
IPR011682 Glyco_hydro_38_C
IPR001394 Peptidase_C19_UCH
SUPFAM
CDD
ProteinModelPortal
H9J9Q3
A0A2W1C0I7
A0A194QBR2
A0A2A4JIT3
A0A2H1V9X6
A0A194QVQ3
+ More
A0A212F922 A0A182SMD5 A0A2J7Q031 F5HL44 A0A182XJ28 A0A182V0I8 A0A182I7T1 A0A1B6LG77 A0A182P1Q3 A0A182LYI4 T1E996 A0A067QF27 A0A182TL51 A0A182YGM6 A0A182W1I7 A0A182IQ03 A0A182K1D7 T1PDX1 A0A182RYM9 T1PDU2 A0A182R0A2 A0A182N5S8 A0A2M4ABQ9 A0A2M4ABS9 W5JPA9 A0A182FPU6 A0A1L8E0R1 A0A0K8TU30 A0A2H8TEF4 A0A1S4EXR8 A0A1B6H4G2 Q17M99 A0A2M4BHA3 A0A2M4BHC5 A0A182H086 A0A182GYQ8 A0A2M3Z470 A0A1I8NSM9 A0A2M3Z419 U5EUA8 D6WC30 B0WQC9 A0A336MLA2 A0A1B6DG24 A0A1B6C744 A0A023EXF3 A0A1Q3FI86 A0A1B0DBG6 A0A2P8XVF2 V5GLS7 A0A1B6K6A7 A0A0A9XJW4 J9K007 A0A2H8TYS0 A0A2S2NBL0 A0A0L0BYF3 B4PRU5 T1I5N5 B4IC50 A0A0R1E5P7 A0A2S2Q9I8 A0A023F4G0 A0A0V0GBL2 A0A1W4UIU7 H9J9Q4 A0A2J7Q043 B4KAX8 A0A3B0KAI4 A0A0Q9WYS2 A0A1W4UH31 B4NBE9 A0A0P4VX09 A0A0M5J924 A0A1J1I538 A0A3B0K3M4 A0A0Q9WT09 A0A3B0JE23 B4G2G6 B4JU20 B4LXG6 A0A0P4W311 A0A0Q9WG72 Q9U1J0 A0A1B0B2N3 B3P850 Q9VB69 A0A0P4W9N3 E1JIZ4 E1JIZ5 W8BCJ7 A0A1A9YSV7 A0A1B0FEH5 Q29AC6 A0A1A9ZZ14
A0A212F922 A0A182SMD5 A0A2J7Q031 F5HL44 A0A182XJ28 A0A182V0I8 A0A182I7T1 A0A1B6LG77 A0A182P1Q3 A0A182LYI4 T1E996 A0A067QF27 A0A182TL51 A0A182YGM6 A0A182W1I7 A0A182IQ03 A0A182K1D7 T1PDX1 A0A182RYM9 T1PDU2 A0A182R0A2 A0A182N5S8 A0A2M4ABQ9 A0A2M4ABS9 W5JPA9 A0A182FPU6 A0A1L8E0R1 A0A0K8TU30 A0A2H8TEF4 A0A1S4EXR8 A0A1B6H4G2 Q17M99 A0A2M4BHA3 A0A2M4BHC5 A0A182H086 A0A182GYQ8 A0A2M3Z470 A0A1I8NSM9 A0A2M3Z419 U5EUA8 D6WC30 B0WQC9 A0A336MLA2 A0A1B6DG24 A0A1B6C744 A0A023EXF3 A0A1Q3FI86 A0A1B0DBG6 A0A2P8XVF2 V5GLS7 A0A1B6K6A7 A0A0A9XJW4 J9K007 A0A2H8TYS0 A0A2S2NBL0 A0A0L0BYF3 B4PRU5 T1I5N5 B4IC50 A0A0R1E5P7 A0A2S2Q9I8 A0A023F4G0 A0A0V0GBL2 A0A1W4UIU7 H9J9Q4 A0A2J7Q043 B4KAX8 A0A3B0KAI4 A0A0Q9WYS2 A0A1W4UH31 B4NBE9 A0A0P4VX09 A0A0M5J924 A0A1J1I538 A0A3B0K3M4 A0A0Q9WT09 A0A3B0JE23 B4G2G6 B4JU20 B4LXG6 A0A0P4W311 A0A0Q9WG72 Q9U1J0 A0A1B0B2N3 B3P850 Q9VB69 A0A0P4W9N3 E1JIZ4 E1JIZ5 W8BCJ7 A0A1A9YSV7 A0A1B0FEH5 Q29AC6 A0A1A9ZZ14
PDB
1GQ2
E-value=1.09102e-71,
Score=684
Ontologies
GO
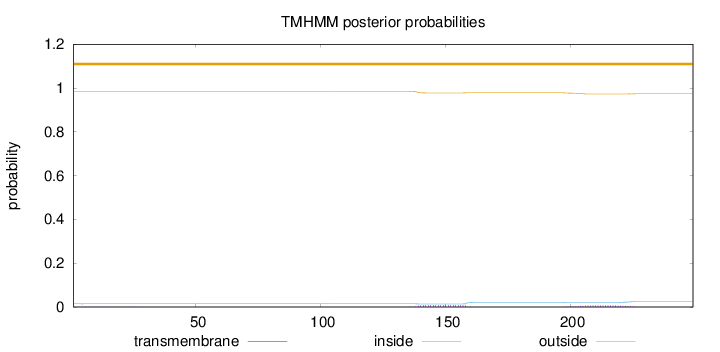
Topology
Length:
249
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.3319
Exp number, first 60 AAs:
0.00534
Total prob of N-in:
0.01463
outside
1 - 249
Population Genetic Test Statistics
Pi
169.951239
Theta
17.659563
Tajima's D
-0.639652
CLR
1.657539
CSRT
0.212739363031848
Interpretation
Uncertain
