Pre Gene Modal
BGIBMGA006243
Annotation
vacuolar_protein_sorting_4_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.602
Sequence
CDS
ATGACATCATCAAATACCTTACAAAAAGCTATTGATCTTGTCACTAAGGCTACTGAAGAGGACAAGAACAAGAACTATGAAGAAGCATTACGTTTATATGAGCACGGTGTAGAGTATTTTCTTCATGCAGTAAAATATGAGGCCCAAGGTGAAAGGGCAAAAGAAAGCATTCGTGCAAAATGCCTTCAATATCTTGATAGAGCTGAAAAATTAAAGGAATATCTCAAAAAAGATCAAAAGAAAAAACCAGTAAAGGATGGGGAATCCAAAAGTGATGATAAGAAGAGTGATAGTGACAGTGATTCTGATGATCCAGAAAAGAAAAAACTACAAGGGAAACTTGAAGGAGCTATTGTGGTAGAGAAACCTCATGTGAAGTGGAGTGATGTAGCTGGTCTAGAAGCCGCTAAGGAAGCATTGAAAGAGGCTGTAATTCTACCAATAAAATTTCCACATTTGTTTACTGGCAAGAGAATACCATGGAAGGGCATCTTATTATTTGGACCCCCTGGTACTGGTAAGTCATACTTGGCAAAGGCTGTGGCTACTGAAGCAAACAATTCAACATTCTTCTCTGTTTCATCATCTGATCTGGTTTCTAAATGGCTGGGTGAGTCGGAGAAGTTGGTAAAGAACTTATTTGACCTTGCTCGACAACATAAACCGAGCATCATCTTTATTGATGAAATAGATTCACTTTGCTCTTCCCGTTCAGACAACGAATCTGAATCGGCTCGTAGGATCAAGACAGAATTCCTTGTTCAAATGCAAGGTGTTGGTAATGATATGGATGGCATTTTAGTTCTTGGTGCCACAAATATACCATGGGTCCTTGATTCTGCTATTCGAAGGCGTTTTGAGAAGCGTATTTACATTGCTCTGCCAGAAGAACATGCCCGTCTTGATATGTTTAAGTTGCATTTAGGCAACACAAGACATCAACTGTCTGAACAAGATATGAAATTATTAGCGGCTAAAAGTGAGGGATATTCCGGTGCGGATATCAGCATTGTTGTCCGTGATGCACTAATGCAACCTGTACGTAAAGTACAATCTGCCACACACTTTAAAAAAATATCTGGTCCGAGCCCAACTGATCCTAATGTTATTGTCAATGACCTTCTGACGCCATGTTCGCCTGGTGACCCAGGAGCTATAGAAATGACCTGGATCGATGTGCCTAGTGACAAACTTGGCGAGCCGCCAGTTACAATGTCAGACATGTTGAGATCTTTAGCTGTTTCGAAGCCTACTGTAAATGATGATGATATGGTAAAATTGAGGAAATTCATGGAAGACTTTGGTCAGGAAGGATAA
Protein
MTSSNTLQKAIDLVTKATEEDKNKNYEEALRLYEHGVEYFLHAVKYEAQGERAKESIRAKCLQYLDRAEKLKEYLKKDQKKKPVKDGESKSDDKKSDSDSDSDDPEKKKLQGKLEGAIVVEKPHVKWSDVAGLEAAKEALKEAVILPIKFPHLFTGKRIPWKGILLFGPPGTGKSYLAKAVATEANNSTFFSVSSSDLVSKWLGESEKLVKNLFDLARQHKPSIIFIDEIDSLCSSRSDNESESARRIKTEFLVQMQGVGNDMDGILVLGATNIPWVLDSAIRRRFEKRIYIALPEEHARLDMFKLHLGNTRHQLSEQDMKLLAAKSEGYSGADISIVVRDALMQPVRKVQSATHFKKISGPSPTDPNVIVNDLLTPCSPGDPGAIEMTWIDVPSDKLGEPPVTMSDMLRSLAVSKPTVNDDDMVKLRKFMEDFGQEG
Summary
Similarity
Belongs to the AAA ATPase family.
Uniprot
D0FH76
A0A194QAH5
A0A194QQ60
A0A0L7LRQ1
S4NP65
A0A212F7F0
+ More
G3LZX7 A0A2W1BY93 A0A2A4JHS1 A0A1B6GRT2 A0A1B6JKV1 A0A1B0CEA3 E0VZT1 A0A2J7REM5 A0A1L8DW85 A0A224XCM5 A0A0V0G4F4 A0A0N9DVI3 D6WFH6 A0A023FCK6 A0A034VKV7 A0A0K8VZI4 A0A0A1WJB3 W8BH79 A0A1A9X821 A0A1B0ALW9 Q17GP3 A0A182J150 R4WDE1 A0A1A9ZLK4 A0A1A9WIK9 D3TM40 A0A182VJ69 A0A1I8NLR4 A0A1A9UU52 A0A1B0EXD1 Q7QFR0 A0A1L8EH31 N6TV25 A0A1B6DX78 A0A182MTG9 A0A023EUM0 A0A0A9YRA5 A0A182R227 A0A067RG64 A0A182VVI2 T1PLV6 A0A1Q3FEB8 A0A182PM57 A0A0L0CCI1 A0A182GN96 A0A182N225 A0A182N143 J9JLV9 B4L2B2 A0A182QVL6 A0A2H8TML2 B3MXW2 B4M6S6 A0A3B0J8P5 A0A2J7REN3 A0A0K8TSC7 Q29H77 B4R6Q7 A0A2S2PDY0 B4I6L5 B3NWZ3 Q9Y162 A0A3L8D966 B4Q2M1 A0A1W4UN90 A0A336MBZ7 B0XJH8 A0A1B6GL33 A0A1J1HHD7 B4NPI4 A0A182QKU6 A0A0M9A6X9 A0A088A3I7 A0A084VIV8 A0A0M3QYW5 A0A154NYG2 F6ZXA3 B2GUK1 K7J635 A0A182XYD4 A0A226NCZ1 E2BQY9 W5M4P7 A0A0L7QSE9 A0A1L8GF31 A0A226PG52 A0A1D5NYD8 W5JUK6 Q66IY7 A0A226EZ97 T1DQY8 A0A182FIV7 A0A2M4AAH7 A0A1V4K1I7
G3LZX7 A0A2W1BY93 A0A2A4JHS1 A0A1B6GRT2 A0A1B6JKV1 A0A1B0CEA3 E0VZT1 A0A2J7REM5 A0A1L8DW85 A0A224XCM5 A0A0V0G4F4 A0A0N9DVI3 D6WFH6 A0A023FCK6 A0A034VKV7 A0A0K8VZI4 A0A0A1WJB3 W8BH79 A0A1A9X821 A0A1B0ALW9 Q17GP3 A0A182J150 R4WDE1 A0A1A9ZLK4 A0A1A9WIK9 D3TM40 A0A182VJ69 A0A1I8NLR4 A0A1A9UU52 A0A1B0EXD1 Q7QFR0 A0A1L8EH31 N6TV25 A0A1B6DX78 A0A182MTG9 A0A023EUM0 A0A0A9YRA5 A0A182R227 A0A067RG64 A0A182VVI2 T1PLV6 A0A1Q3FEB8 A0A182PM57 A0A0L0CCI1 A0A182GN96 A0A182N225 A0A182N143 J9JLV9 B4L2B2 A0A182QVL6 A0A2H8TML2 B3MXW2 B4M6S6 A0A3B0J8P5 A0A2J7REN3 A0A0K8TSC7 Q29H77 B4R6Q7 A0A2S2PDY0 B4I6L5 B3NWZ3 Q9Y162 A0A3L8D966 B4Q2M1 A0A1W4UN90 A0A336MBZ7 B0XJH8 A0A1B6GL33 A0A1J1HHD7 B4NPI4 A0A182QKU6 A0A0M9A6X9 A0A088A3I7 A0A084VIV8 A0A0M3QYW5 A0A154NYG2 F6ZXA3 B2GUK1 K7J635 A0A182XYD4 A0A226NCZ1 E2BQY9 W5M4P7 A0A0L7QSE9 A0A1L8GF31 A0A226PG52 A0A1D5NYD8 W5JUK6 Q66IY7 A0A226EZ97 T1DQY8 A0A182FIV7 A0A2M4AAH7 A0A1V4K1I7
Pubmed
19121390
23053938
26354079
26227816
23622113
22118469
+ More
22072775 28756777 20566863 18362917 19820115 25474469 25348373 25830018 24495485 17510324 23691247 20353571 12364791 14747013 17210077 23537049 24945155 25401762 26823975 24845553 25315136 26108605 26483478 17994087 26369729 15632085 23185243 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 30249741 17550304 24438588 20431018 20075255 25244985 20798317 27762356 24621616 15592404 20920257 23761445
22072775 28756777 20566863 18362917 19820115 25474469 25348373 25830018 24495485 17510324 23691247 20353571 12364791 14747013 17210077 23537049 24945155 25401762 26823975 24845553 25315136 26108605 26483478 17994087 26369729 15632085 23185243 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 30249741 17550304 24438588 20431018 20075255 25244985 20798317 27762356 24621616 15592404 20920257 23761445
EMBL
BABH01034002
GQ995504
ACX69978.1
KQ459249
KPJ02434.1
KQ461181
+ More
KPJ07632.1 JTDY01000289 KOB77876.1 GAIX01011969 JAA80591.1 AGBW02009892 OWR49629.1 JN584642 AEO21929.1 KZ149899 PZC78625.1 NWSH01001337 PCG71625.1 GECZ01004635 JAS65134.1 GECU01016213 GECU01007800 JAS91493.1 JAS99906.1 AJWK01008702 DS235854 EEB18887.1 NEVH01004959 PNF39285.1 GFDF01003390 JAV10694.1 GFTR01006240 JAW10186.1 GECL01003180 JAP02944.1 KT448842 ALF12302.1 KQ971327 EFA00923.1 GBBI01000189 JAC18523.1 GAKP01016240 JAC42712.1 GDHF01009875 GDHF01008070 JAI42439.1 JAI44244.1 GBXI01015546 JAC98745.1 GAMC01008452 GAMC01008450 GAMC01008448 JAB98107.1 JXJN01000170 CH477257 EAT45823.1 EAT45824.1 AK417598 BAN20813.1 CCAG010000401 EZ422492 ADD18768.1 AJVK01001010 AJVK01001011 AAAB01008846 EAA06410.2 GFDG01000835 JAV17964.1 APGK01053679 KB741231 KB632387 ENN72251.1 ERL94352.1 GEDC01012581 GEDC01007027 JAS24717.1 JAS30271.1 AXCM01014345 GAPW01001439 JAC12159.1 GBHO01008850 GDHC01015818 JAG34754.1 JAQ02811.1 KK852482 KDR22866.1 KA649366 KA649767 AFP64396.1 GFDL01009187 JAV25858.1 JRES01000575 KNC30143.1 JXUM01076042 JXUM01076043 KQ562920 KXJ74841.1 ABLF02021602 CH933810 EDW07773.1 AXCN02002167 GFXV01003464 MBW15269.1 CH902630 EDV38577.1 CH940653 EDW62493.1 OUUW01000003 SPP78557.1 PNF39286.1 GDAI01000339 JAI17264.1 CH379064 EAL31881.1 KRT06635.1 CM000366 EDX18256.1 GGMR01015044 MBY27663.1 CH480823 EDW55963.1 CH954180 EDV46540.1 AF145606 AE014298 AAD38581.1 AAF48783.1 AHN59866.1 QOIP01000011 RLU16861.1 CM000162 EDX02662.1 UFQT01000833 SSX27470.1 DS233521 EDS30381.1 GECZ01006605 JAS63164.1 CVRI01000004 CRK87440.1 CH964291 EDW86424.1 AXCN02002351 KQ435724 KOX78477.1 ATLV01013411 KE524855 KFB37902.1 CP012528 ALC48393.1 KQ434783 KZC04716.1 AAMC01141857 BC166312 AAI66312.1 MCFN01000104 OXB65089.1 GL449825 EFN81892.1 AHAT01010607 AHAT01010608 KQ414758 KOC61479.1 CM004473 OCT82385.1 AWGT02000084 OXB78367.1 AADN05000558 ADMH02000095 ETN67836.1 BC081138 CM004472 AAH81138.1 OCT84476.1 LNIX01000001 OXA62902.1 GAMD01000885 JAB00706.1 GGFK01004458 MBW37779.1 LSYS01005191 OPJ78296.1
KPJ07632.1 JTDY01000289 KOB77876.1 GAIX01011969 JAA80591.1 AGBW02009892 OWR49629.1 JN584642 AEO21929.1 KZ149899 PZC78625.1 NWSH01001337 PCG71625.1 GECZ01004635 JAS65134.1 GECU01016213 GECU01007800 JAS91493.1 JAS99906.1 AJWK01008702 DS235854 EEB18887.1 NEVH01004959 PNF39285.1 GFDF01003390 JAV10694.1 GFTR01006240 JAW10186.1 GECL01003180 JAP02944.1 KT448842 ALF12302.1 KQ971327 EFA00923.1 GBBI01000189 JAC18523.1 GAKP01016240 JAC42712.1 GDHF01009875 GDHF01008070 JAI42439.1 JAI44244.1 GBXI01015546 JAC98745.1 GAMC01008452 GAMC01008450 GAMC01008448 JAB98107.1 JXJN01000170 CH477257 EAT45823.1 EAT45824.1 AK417598 BAN20813.1 CCAG010000401 EZ422492 ADD18768.1 AJVK01001010 AJVK01001011 AAAB01008846 EAA06410.2 GFDG01000835 JAV17964.1 APGK01053679 KB741231 KB632387 ENN72251.1 ERL94352.1 GEDC01012581 GEDC01007027 JAS24717.1 JAS30271.1 AXCM01014345 GAPW01001439 JAC12159.1 GBHO01008850 GDHC01015818 JAG34754.1 JAQ02811.1 KK852482 KDR22866.1 KA649366 KA649767 AFP64396.1 GFDL01009187 JAV25858.1 JRES01000575 KNC30143.1 JXUM01076042 JXUM01076043 KQ562920 KXJ74841.1 ABLF02021602 CH933810 EDW07773.1 AXCN02002167 GFXV01003464 MBW15269.1 CH902630 EDV38577.1 CH940653 EDW62493.1 OUUW01000003 SPP78557.1 PNF39286.1 GDAI01000339 JAI17264.1 CH379064 EAL31881.1 KRT06635.1 CM000366 EDX18256.1 GGMR01015044 MBY27663.1 CH480823 EDW55963.1 CH954180 EDV46540.1 AF145606 AE014298 AAD38581.1 AAF48783.1 AHN59866.1 QOIP01000011 RLU16861.1 CM000162 EDX02662.1 UFQT01000833 SSX27470.1 DS233521 EDS30381.1 GECZ01006605 JAS63164.1 CVRI01000004 CRK87440.1 CH964291 EDW86424.1 AXCN02002351 KQ435724 KOX78477.1 ATLV01013411 KE524855 KFB37902.1 CP012528 ALC48393.1 KQ434783 KZC04716.1 AAMC01141857 BC166312 AAI66312.1 MCFN01000104 OXB65089.1 GL449825 EFN81892.1 AHAT01010607 AHAT01010608 KQ414758 KOC61479.1 CM004473 OCT82385.1 AWGT02000084 OXB78367.1 AADN05000558 ADMH02000095 ETN67836.1 BC081138 CM004472 AAH81138.1 OCT84476.1 LNIX01000001 OXA62902.1 GAMD01000885 JAB00706.1 GGFK01004458 MBW37779.1 LSYS01005191 OPJ78296.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000037510
UP000007151
UP000218220
+ More
UP000092461 UP000009046 UP000235965 UP000007266 UP000092443 UP000092460 UP000008820 UP000075880 UP000092445 UP000091820 UP000092444 UP000075903 UP000095300 UP000078200 UP000092462 UP000007062 UP000019118 UP000030742 UP000075883 UP000075900 UP000027135 UP000075920 UP000095301 UP000075885 UP000037069 UP000069940 UP000249989 UP000075884 UP000007819 UP000009192 UP000075886 UP000007801 UP000008792 UP000268350 UP000001819 UP000000304 UP000001292 UP000008711 UP000000803 UP000279307 UP000002282 UP000192221 UP000002320 UP000183832 UP000007798 UP000053105 UP000005203 UP000030765 UP000092553 UP000076502 UP000008143 UP000002358 UP000076408 UP000198323 UP000008237 UP000018468 UP000053825 UP000186698 UP000198419 UP000000539 UP000000673 UP000198287 UP000069272 UP000190648
UP000092461 UP000009046 UP000235965 UP000007266 UP000092443 UP000092460 UP000008820 UP000075880 UP000092445 UP000091820 UP000092444 UP000075903 UP000095300 UP000078200 UP000092462 UP000007062 UP000019118 UP000030742 UP000075883 UP000075900 UP000027135 UP000075920 UP000095301 UP000075885 UP000037069 UP000069940 UP000249989 UP000075884 UP000007819 UP000009192 UP000075886 UP000007801 UP000008792 UP000268350 UP000001819 UP000000304 UP000001292 UP000008711 UP000000803 UP000279307 UP000002282 UP000192221 UP000002320 UP000183832 UP000007798 UP000053105 UP000005203 UP000030765 UP000092553 UP000076502 UP000008143 UP000002358 UP000076408 UP000198323 UP000008237 UP000018468 UP000053825 UP000186698 UP000198419 UP000000539 UP000000673 UP000198287 UP000069272 UP000190648
Interpro
Gene 3D
CDD
ProteinModelPortal
D0FH76
A0A194QAH5
A0A194QQ60
A0A0L7LRQ1
S4NP65
A0A212F7F0
+ More
G3LZX7 A0A2W1BY93 A0A2A4JHS1 A0A1B6GRT2 A0A1B6JKV1 A0A1B0CEA3 E0VZT1 A0A2J7REM5 A0A1L8DW85 A0A224XCM5 A0A0V0G4F4 A0A0N9DVI3 D6WFH6 A0A023FCK6 A0A034VKV7 A0A0K8VZI4 A0A0A1WJB3 W8BH79 A0A1A9X821 A0A1B0ALW9 Q17GP3 A0A182J150 R4WDE1 A0A1A9ZLK4 A0A1A9WIK9 D3TM40 A0A182VJ69 A0A1I8NLR4 A0A1A9UU52 A0A1B0EXD1 Q7QFR0 A0A1L8EH31 N6TV25 A0A1B6DX78 A0A182MTG9 A0A023EUM0 A0A0A9YRA5 A0A182R227 A0A067RG64 A0A182VVI2 T1PLV6 A0A1Q3FEB8 A0A182PM57 A0A0L0CCI1 A0A182GN96 A0A182N225 A0A182N143 J9JLV9 B4L2B2 A0A182QVL6 A0A2H8TML2 B3MXW2 B4M6S6 A0A3B0J8P5 A0A2J7REN3 A0A0K8TSC7 Q29H77 B4R6Q7 A0A2S2PDY0 B4I6L5 B3NWZ3 Q9Y162 A0A3L8D966 B4Q2M1 A0A1W4UN90 A0A336MBZ7 B0XJH8 A0A1B6GL33 A0A1J1HHD7 B4NPI4 A0A182QKU6 A0A0M9A6X9 A0A088A3I7 A0A084VIV8 A0A0M3QYW5 A0A154NYG2 F6ZXA3 B2GUK1 K7J635 A0A182XYD4 A0A226NCZ1 E2BQY9 W5M4P7 A0A0L7QSE9 A0A1L8GF31 A0A226PG52 A0A1D5NYD8 W5JUK6 Q66IY7 A0A226EZ97 T1DQY8 A0A182FIV7 A0A2M4AAH7 A0A1V4K1I7
G3LZX7 A0A2W1BY93 A0A2A4JHS1 A0A1B6GRT2 A0A1B6JKV1 A0A1B0CEA3 E0VZT1 A0A2J7REM5 A0A1L8DW85 A0A224XCM5 A0A0V0G4F4 A0A0N9DVI3 D6WFH6 A0A023FCK6 A0A034VKV7 A0A0K8VZI4 A0A0A1WJB3 W8BH79 A0A1A9X821 A0A1B0ALW9 Q17GP3 A0A182J150 R4WDE1 A0A1A9ZLK4 A0A1A9WIK9 D3TM40 A0A182VJ69 A0A1I8NLR4 A0A1A9UU52 A0A1B0EXD1 Q7QFR0 A0A1L8EH31 N6TV25 A0A1B6DX78 A0A182MTG9 A0A023EUM0 A0A0A9YRA5 A0A182R227 A0A067RG64 A0A182VVI2 T1PLV6 A0A1Q3FEB8 A0A182PM57 A0A0L0CCI1 A0A182GN96 A0A182N225 A0A182N143 J9JLV9 B4L2B2 A0A182QVL6 A0A2H8TML2 B3MXW2 B4M6S6 A0A3B0J8P5 A0A2J7REN3 A0A0K8TSC7 Q29H77 B4R6Q7 A0A2S2PDY0 B4I6L5 B3NWZ3 Q9Y162 A0A3L8D966 B4Q2M1 A0A1W4UN90 A0A336MBZ7 B0XJH8 A0A1B6GL33 A0A1J1HHD7 B4NPI4 A0A182QKU6 A0A0M9A6X9 A0A088A3I7 A0A084VIV8 A0A0M3QYW5 A0A154NYG2 F6ZXA3 B2GUK1 K7J635 A0A182XYD4 A0A226NCZ1 E2BQY9 W5M4P7 A0A0L7QSE9 A0A1L8GF31 A0A226PG52 A0A1D5NYD8 W5JUK6 Q66IY7 A0A226EZ97 T1DQY8 A0A182FIV7 A0A2M4AAH7 A0A1V4K1I7
PDB
2ZAO
E-value=1.80415e-176,
Score=1590
Ontologies
GO
GO:0005524
GO:0008568
GO:0005634
GO:0007033
GO:0016887
GO:0046983
GO:0016322
GO:0043162
GO:0045470
GO:0044130
GO:0045197
GO:0097352
GO:0045463
GO:0030036
GO:0045742
GO:1990621
GO:0005768
GO:0048471
GO:0048524
GO:0061738
GO:0036258
GO:0007165
GO:0008270
GO:0016020
GO:0004352
GO:0000287
GO:0003723
GO:0006270
GO:0042555
GO:0048384
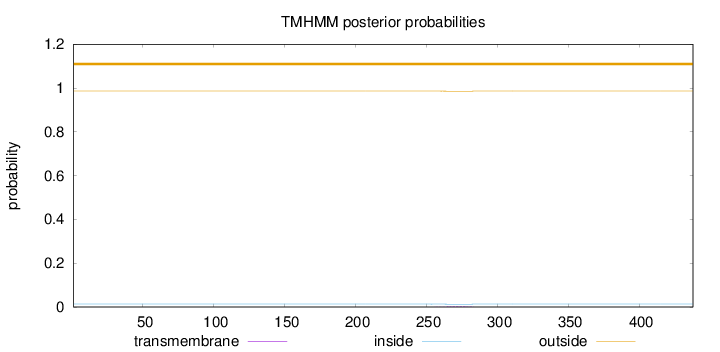
Topology
Length:
438
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.05416
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01343
outside
1 - 438
Population Genetic Test Statistics
Pi
148.609917
Theta
162.927444
Tajima's D
-1.155132
CLR
0.035033
CSRT
0.111594420278986
Interpretation
Uncertain
