Gene
KWMTBOMO03510
Pre Gene Modal
BGIBMGA006244
Annotation
putative_Protein_toll_[Operophtera_brumata]
Location in the cell
Nuclear Reliability : 2.054
Sequence
CDS
ATGGGTCGCAAAATGCTGCGTTATTATTTGGCAGCTGTGCTACTTCAAGTGGTGGCCGCGACAAACATTTCATGTCCGACTGACGCCAATTGTACTTGTGGCGGTACCTTGGCTATCGAGCTTGACTGCAATATCGATGGAAGCGTTGTGAACATAAATCTAGTACCGGAGATTTACATCAATGTCAAATGTGAGAATGCAACGGCACTCGATTATAGCAAACTACCGAGATGTACAAATATAACTGATTCATTCCAATCCGTTAGCTTCAAGGACTGTCCATTACCGGAGACTTCTTTTAAAGATGTTTTAGACAACATGGGCGTCACTAAAACTATGGCACTAATCTTCCAGAATGCCAAGAATCTATCGGGATACTTAGATCGAAAACATTTCGCTGGATTACAAGATTTAACCAAACTATTATTGTCAATTAACGGAGTTATTCATCTACCAGACGACGCTTTTGTTGACATCAACAAACTTACTTGGTTGAACATAAGATCGAAAACTATATATCTGTCTGAGGAGCTATTTAAGCCCTTGGAAAGGCTGGAAACTTTAGAAATTAGTCACAACCACATGACTAATATAACTTCAAACTTGTTTTCTCATCTATCCTTCTTGAGGAAACTTTCGTTATGGCAGAGTAATGTTACCTGGTTTTCCCAGGACTTTTTCAACGGCGTAGACGTTCTCGAAGAATTAGATTTAAGTTCTAATGGACTTAACAAACTTCCGGCGACAATATTTAAGCCTCTCAAGAAATTAAAGAAACTCACTTTATTTTCGAACAAATTTTCATCTTTACCGAAGAACTTATTCATCGGCAACAACGAACTGGAAACTGTAGTTATTTTAAACAATGATGTTAAATTAAGGGTACTGCCGGAACGTTTGTTTGGAAATTTGCCCAATTTGAAGCAGGTTTACGTTCAACGAAGCGGTATCGAGGTCGTGCCACGCGACACGTTCAAGAACTCACCGGCAATTACCAACATATCGTTAGCGTTCAACGATTTAAAACGCCTCCCGGAGGAAGTGTTCAACAATCAGATCAATTTATTAAATTTAGATCTCAGTCACAACAAGATCTTTGATTTGGCAACGAGAACGTTTTCAACGTTGGTGCGACTCGAGATATTGAACTTGTGTAATAATCAACTTGTGTTTATTGATGGATCACTATTCTCTTCTTTGTTAAGCCTTATCGACCTGAATATAGAGAATAATAATTTAAAGATTATTACGTCGGATTTGTTTCGGAACAACAAACAGAGGATGTCTATTTCGTTGGCAAACAATAATCTCGACTTCGAGCAGACAGTGTACGCGAACAACTGTTTGTCTCTGGAACCGAGATCGCCATTTGCAAATACTTACAACTTGAAGCTATTGAATCTGAGTCATAACGCGTTCAGGATCAGTTTCGTGGACTGGTGGATTAATGGTCATGAAACCGTTGATGTCAGTCACAATGCAATTAAAACTCTCGTGGCTTACAATGAGTATTCGGATTTATACGAGGGTGTTACCAGAAAGCCACTCAAAGAACTGTGGGTTTTCAATAATCCCATTCAATGTGTTTGCAACAATTACATGTTCGTTGATCTTCTTCAAGACGTAACAAACCGTAAGATTATGGATGCACGCTTGTTCCGTTGCCCGAAATGGCAAATAACATTCTGCAGTATTCGTGTTCTAACGATTTCGGCCATTATTTTGGGCTTGTGTGTGATGTACGCGGTCATCACCATGGCGTATTATATATACAAGTGTCGAGCGTACGAGACTCTTAAGTGGAAGCTGTCTTTACTGTCGAAAAAGGGCATACGGATTATCGAACAAGAATTAATAATAAGATACTCAGAAAAGGACGAAGAGTTTGTAATGAAAGAGATACTGCCTACGTTAAAAAATGAGAAAAATTTGAAAGTGTACACAAACGCGATCAGGTGTCCCAGCGATATCAGCCCGGACAACCTGAAGACCTGCGTCAAGGATGCTTACAAGTACACCACCTTAATAGTTTTCTCCCCAAATTATTTGACGACAGCCTACAGCAACGTGGACATCAAGAAAATACACAGCGAAATGTTGAAGGCTGAATACACTGTGTACGTTTTTGTGGACATCGGCCCTGAGAACTTCCTGTACGTTTTCCTGAAGAGCCAACGCGATACGAACACAATGGTGGTTTGGGGCGAAGCTAATTTTTGGTTAAAGTTCCTGGTGACCTTGGCTCAATACGACATTACCTATCCTGATCTTAACAAGATTTCCGATAAAATAACTTTTGCCGAAGTCGAGGAGAATGATAAAAAGAATTGTCACCTCTGTTACTTAGGGACTCACCGTTGCCACAGCTGCTGCTACGGTATTATTCTAGACCAGAGTCTAGTTTGA
Protein
MGRKMLRYYLAAVLLQVVAATNISCPTDANCTCGGTLAIELDCNIDGSVVNINLVPEIYINVKCENATALDYSKLPRCTNITDSFQSVSFKDCPLPETSFKDVLDNMGVTKTMALIFQNAKNLSGYLDRKHFAGLQDLTKLLLSINGVIHLPDDAFVDINKLTWLNIRSKTIYLSEELFKPLERLETLEISHNHMTNITSNLFSHLSFLRKLSLWQSNVTWFSQDFFNGVDVLEELDLSSNGLNKLPATIFKPLKKLKKLTLFSNKFSSLPKNLFIGNNELETVVILNNDVKLRVLPERLFGNLPNLKQVYVQRSGIEVVPRDTFKNSPAITNISLAFNDLKRLPEEVFNNQINLLNLDLSHNKIFDLATRTFSTLVRLEILNLCNNQLVFIDGSLFSSLLSLIDLNIENNNLKIITSDLFRNNKQRMSISLANNNLDFEQTVYANNCLSLEPRSPFANTYNLKLLNLSHNAFRISFVDWWINGHETVDVSHNAIKTLVAYNEYSDLYEGVTRKPLKELWVFNNPIQCVCNNYMFVDLLQDVTNRKIMDARLFRCPKWQITFCSIRVLTISAIILGLCVMYAVITMAYYIYKCRAYETLKWKLSLLSKKGIRIIEQELIIRYSEKDEEFVMKEILPTLKNEKNLKVYTNAIRCPSDISPDNLKTCVKDAYKYTTLIVFSPNYLTTAYSNVDIKKIHSEMLKAEYTVYVFVDIGPENFLYVFLKSQRDTNTMVVWGEANFWLKFLVTLAQYDITYPDLNKISDKITFAEVEENDKKNCHLCYLGTHRCHSCCYGIILDQSLV
Summary
Similarity
Belongs to the Toll-like receptor family.
Uniprot
H9J9Q1
A0A2W1BU69
A0A2A4JPC4
A0A2H1WSD4
A0A0L7LDR6
A0A212FFN9
+ More
A0A194QXR0 A0A1I8PSW5 A0A067R3H7 A0A2J7QHR7 A0A2J7QHL1 A0A1A9ZL81 A0A1A9VTQ4 A0A1B0GER6 A0A1A9XZE1 A0A182Y4Y2 A0A182M8X8 A0A182GPV4 A0A2C9GWS9 A0A2C9H6H6 A0A182QPS8 A0A182PVM9 Q171J8 A0A182KC65 Q8MQU8 Q8MQU7 A0A182UFK6 A0A154NYN7 A0A0D3QDC7 Q8WRE5 A0A0D3QDD7 A0A0D3QDL3 A0A0D3QDL8 A0A0D3QCR2 A0A0D3QCQ2 A0A0D3QD03 A0A182IDR8 A0A0D3QCQ7 A0A182XNF0 Q17F73 A0A2C9GH57 E9IK94 A0A1A9W5W9 A0A084WPQ0 A0A195CSW9 A0A336MUL7
A0A194QXR0 A0A1I8PSW5 A0A067R3H7 A0A2J7QHR7 A0A2J7QHL1 A0A1A9ZL81 A0A1A9VTQ4 A0A1B0GER6 A0A1A9XZE1 A0A182Y4Y2 A0A182M8X8 A0A182GPV4 A0A2C9GWS9 A0A2C9H6H6 A0A182QPS8 A0A182PVM9 Q171J8 A0A182KC65 Q8MQU8 Q8MQU7 A0A182UFK6 A0A154NYN7 A0A0D3QDC7 Q8WRE5 A0A0D3QDD7 A0A0D3QDL3 A0A0D3QDL8 A0A0D3QCR2 A0A0D3QCQ2 A0A0D3QD03 A0A182IDR8 A0A0D3QCQ7 A0A182XNF0 Q17F73 A0A2C9GH57 E9IK94 A0A1A9W5W9 A0A084WPQ0 A0A195CSW9 A0A336MUL7
Pubmed
EMBL
BABH01033973
BABH01033974
KZ149899
PZC78622.1
NWSH01000850
PCG73851.1
+ More
ODYU01010702 SOQ55980.1 JTDY01001536 KOB73622.1 AGBW02008793 OWR52546.1 KQ461155 KPJ08366.1 KK852931 KDR13629.1 NEVH01013973 PNF28083.1 PNF28085.1 CCAG010012576 CCAG010012577 CCAG010012578 CCAG010012579 AXCM01000253 JXUM01001876 KQ560129 KXJ84298.1 AXCN02002028 CH477452 EAT40676.1 AY124196 AAM97775.1 AY124197 AAM97776.1 KQ434773 KZC03990.1 KP274533 KP274534 KP274535 KP274536 KP274537 KP274538 KP274539 KP274540 KP274541 KP274544 KP274551 AJC98679.1 AJC98680.1 AJC98681.1 AJC98682.1 AJC98683.1 AJC98684.1 AJC98685.1 AJC98686.1 AJC98687.1 AJC98690.1 AJC98697.1 AF444780 AAAB01008811 AAL37901.1 EAA45376.1 KP274549 AJC98695.1 KP274545 KP274548 KP274550 AJC98691.1 AJC98694.1 AJC98696.1 KP274553 AJC98699.1 KP274552 AJC98698.1 KP274542 AJC98688.1 KP274543 KP274546 AJC98689.1 AJC98692.1 APCN01004779 APCN01004780 KP274547 AJC98693.1 CH477273 EAT45212.2 GL763924 EFZ19053.1 ATLV01025119 KE525369 KFB52194.1 KQ977304 KYN03798.1 UFQS01002797 UFQT01002797 SSX14685.1 SSX34082.1
ODYU01010702 SOQ55980.1 JTDY01001536 KOB73622.1 AGBW02008793 OWR52546.1 KQ461155 KPJ08366.1 KK852931 KDR13629.1 NEVH01013973 PNF28083.1 PNF28085.1 CCAG010012576 CCAG010012577 CCAG010012578 CCAG010012579 AXCM01000253 JXUM01001876 KQ560129 KXJ84298.1 AXCN02002028 CH477452 EAT40676.1 AY124196 AAM97775.1 AY124197 AAM97776.1 KQ434773 KZC03990.1 KP274533 KP274534 KP274535 KP274536 KP274537 KP274538 KP274539 KP274540 KP274541 KP274544 KP274551 AJC98679.1 AJC98680.1 AJC98681.1 AJC98682.1 AJC98683.1 AJC98684.1 AJC98685.1 AJC98686.1 AJC98687.1 AJC98690.1 AJC98697.1 AF444780 AAAB01008811 AAL37901.1 EAA45376.1 KP274549 AJC98695.1 KP274545 KP274548 KP274550 AJC98691.1 AJC98694.1 AJC98696.1 KP274553 AJC98699.1 KP274552 AJC98698.1 KP274542 AJC98688.1 KP274543 KP274546 AJC98689.1 AJC98692.1 APCN01004779 APCN01004780 KP274547 AJC98693.1 CH477273 EAT45212.2 GL763924 EFZ19053.1 ATLV01025119 KE525369 KFB52194.1 KQ977304 KYN03798.1 UFQS01002797 UFQT01002797 SSX14685.1 SSX34082.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000007151
UP000053240
UP000095300
+ More
UP000027135 UP000235965 UP000092445 UP000078200 UP000092444 UP000092443 UP000076408 UP000075883 UP000069940 UP000249989 UP000075900 UP000075920 UP000075886 UP000075885 UP000008820 UP000075881 UP000075902 UP000076502 UP000007062 UP000075903 UP000075840 UP000076407 UP000069272 UP000091820 UP000030765 UP000078542
UP000027135 UP000235965 UP000092445 UP000078200 UP000092444 UP000092443 UP000076408 UP000075883 UP000069940 UP000249989 UP000075900 UP000075920 UP000075886 UP000075885 UP000008820 UP000075881 UP000075902 UP000076502 UP000007062 UP000075903 UP000075840 UP000076407 UP000069272 UP000091820 UP000030765 UP000078542
PRIDE
Pfam
Interpro
SUPFAM
SSF52200
SSF52200
Gene 3D
ProteinModelPortal
H9J9Q1
A0A2W1BU69
A0A2A4JPC4
A0A2H1WSD4
A0A0L7LDR6
A0A212FFN9
+ More
A0A194QXR0 A0A1I8PSW5 A0A067R3H7 A0A2J7QHR7 A0A2J7QHL1 A0A1A9ZL81 A0A1A9VTQ4 A0A1B0GER6 A0A1A9XZE1 A0A182Y4Y2 A0A182M8X8 A0A182GPV4 A0A2C9GWS9 A0A2C9H6H6 A0A182QPS8 A0A182PVM9 Q171J8 A0A182KC65 Q8MQU8 Q8MQU7 A0A182UFK6 A0A154NYN7 A0A0D3QDC7 Q8WRE5 A0A0D3QDD7 A0A0D3QDL3 A0A0D3QDL8 A0A0D3QCR2 A0A0D3QCQ2 A0A0D3QD03 A0A182IDR8 A0A0D3QCQ7 A0A182XNF0 Q17F73 A0A2C9GH57 E9IK94 A0A1A9W5W9 A0A084WPQ0 A0A195CSW9 A0A336MUL7
A0A194QXR0 A0A1I8PSW5 A0A067R3H7 A0A2J7QHR7 A0A2J7QHL1 A0A1A9ZL81 A0A1A9VTQ4 A0A1B0GER6 A0A1A9XZE1 A0A182Y4Y2 A0A182M8X8 A0A182GPV4 A0A2C9GWS9 A0A2C9H6H6 A0A182QPS8 A0A182PVM9 Q171J8 A0A182KC65 Q8MQU8 Q8MQU7 A0A182UFK6 A0A154NYN7 A0A0D3QDC7 Q8WRE5 A0A0D3QDD7 A0A0D3QDL3 A0A0D3QDL8 A0A0D3QCR2 A0A0D3QCQ2 A0A0D3QD03 A0A182IDR8 A0A0D3QCQ7 A0A182XNF0 Q17F73 A0A2C9GH57 E9IK94 A0A1A9W5W9 A0A084WPQ0 A0A195CSW9 A0A336MUL7
PDB
4LXS
E-value=2.41177e-46,
Score=471
Ontologies
GO
Topology
SignalP
Position: 1 - 20,
Likelihood: 0.990706
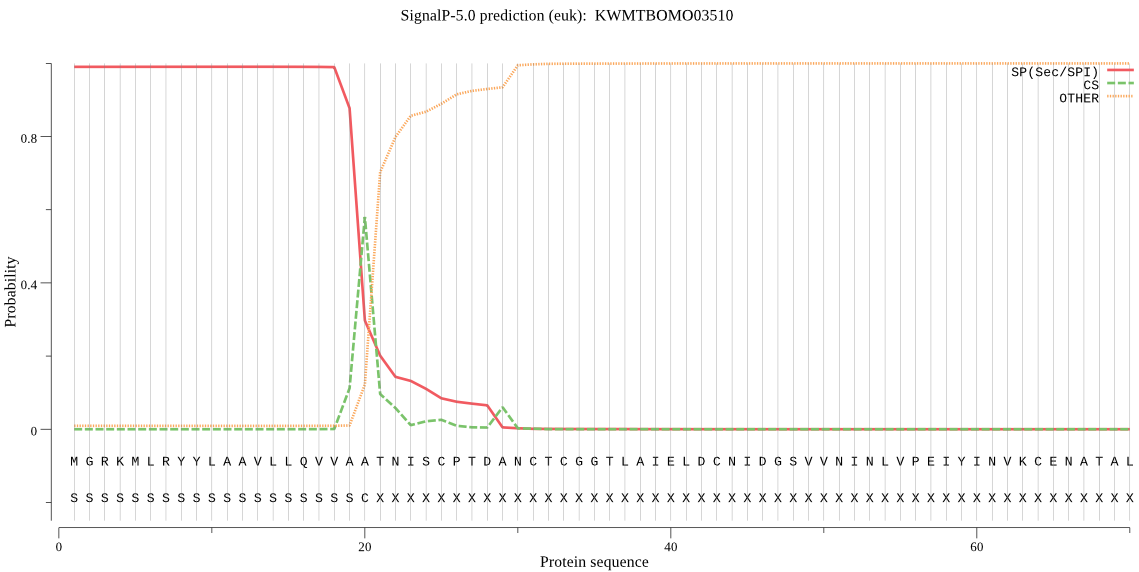
Length:
801
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
27.22176
Exp number, first 60 AAs:
0.3185
Total prob of N-in:
0.03247
outside
1 - 568
TMhelix
569 - 591
inside
592 - 801

Population Genetic Test Statistics
Pi
192.936444
Theta
157.594721
Tajima's D
0.045479
CLR
1.034891
CSRT
0.387230638468077
Interpretation
Uncertain
