Gene
KWMTBOMO03507
Annotation
endonuclease_and_reverse_transcriptase-like_protein_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.299 Nuclear Reliability : 1.162
Sequence
CDS
ATGACCATTAACGCTGGCGTTCCACAAGGTTCGGTGCTCTCCTCCACGCTTTTCATCCTGTATATCAATGACATGCTGTCTATTGATGGCATGCATTGTTATGCGGATGACAGCATGGGGGATGCGCGATATATCGGCCATCAGAGTCTCTCTCGGAGCGTGGTGCAAGAGAAACGATCAAAACTTGTGTCTGAAGTGGAGAACTCTCTGGGGCGAGTCTCCGAATGGGATGAATTGAGCTTGGTTCAATTCAACCCGATAAAGACACAAGTTTGCGCGTTCACTGCGAAGAAGGACCCCTTTGTCATGGCGCCGCAATTCCAAGGAGTATCCCTAGAAACTTCCGGGAGTATCGGGATACTTGGGGTCGACATTTCGAGCGATGTCCAATTTCGGAGTCATTTGGAAGGCAAAGCCAAGTTGGCGTCCAAAATGCTGGGAGTCCTCAACAGAGCAAAGCGGTACTTCACGCCTGGACAAAGACTTTTGCTCTATAAAGCACAAGTCCGGCCTCGCGTGGAGTACTGCTCCCATCTTTAG
Protein
MTINAGVPQGSVLSSTLFILYINDMLSIDGMHCYADDSMGDARYIGHQSLSRSVVQEKRSKLVSEVENSLGRVSEWDELSLVQFNPIKTQVCAFTAKKDPFVMAPQFQGVSLETSGSIGILGVDISSDVQFRSHLEGKAKLASKMLGVLNRAKRYFTPGQRLLLYKAQVRPRVEYCSHL
Summary
Uniprot
Q6UV17
B9WPT7
A0A2H1VFZ3
A0A0P4VXU0
A0A3D5S1K1
A0A1E1XGQ7
+ More
A0A131Y4I8 A0A147BU27 A0A131XU84 A0A2H9T2H3 W4XNV3 A0A1G4M5I2 A0A1E1XNL1 A0A147BQ97 A0A2H1VC59 A0A090XE26 A0A147BLD0 A0A147BKN8 T2M4I6 A0A1E1X2T1 A0A0P4VTV9 A0A2H9T2R7 A0A147BBC3 W4YCB2 A0A2B4RZI8 A0A131Y4Q4 A0A2H9T2Z5 A0A2B4SAL9 A0A147BNQ9 A0A2B4SJF4 A0A147BJQ0 U5EU32 A0A3B3I8F1 A0A2B4S341 A0A2S2PKE0 A0A131XQS8 L7MAD0 A0A2B4RF95 A0A147BB29 A0A1E1X2P5 A0A3B3HI99 A0A0P4VX79 A0A131XQR9 A0A147BJ69 A0A147BBR8 A0A1E1XGC7 A0A2H9T2V7 A0A0K8RPQ6 A0A131Y4G6 A0A131XV56 W4ZFN2 A0A131XRE4 A0A2G8JHC3 A0A147BL45 U5EQW9 A0A2H9T462 A0A2G8LLY0 A0A2H9T2U6 U5EQG0 A0A2G8KET5 A0A3B1IHU1 A0A2B4RTL1 V5H8T0 A0A147BID3 A0A3Q0IWM1 A0A2H9T361 A0A2B4SNL1 A0A147BI04 V5GPT7 A0A2H9T4F1 A0A2H9T3F5 A0A1E1XNX8 A0A147BLD9 W4Y3F1 A0A131XWB2 A0A146PGB4 A0A147BC70 A0A293LCV4 A0A147BK71 A0A146T2W5 A0A146QVS2 A0A182GL77 A0A131XWC9 A0A0P4VPK5 A0A3B3HKX5 A0A131XKD2 A0A0X3PQL9 A0A0X3PL94 A0A147BM11 Q4QQE6 T1DG92 A0A147BNC9 A0A3B1IHX1
A0A131Y4I8 A0A147BU27 A0A131XU84 A0A2H9T2H3 W4XNV3 A0A1G4M5I2 A0A1E1XNL1 A0A147BQ97 A0A2H1VC59 A0A090XE26 A0A147BLD0 A0A147BKN8 T2M4I6 A0A1E1X2T1 A0A0P4VTV9 A0A2H9T2R7 A0A147BBC3 W4YCB2 A0A2B4RZI8 A0A131Y4Q4 A0A2H9T2Z5 A0A2B4SAL9 A0A147BNQ9 A0A2B4SJF4 A0A147BJQ0 U5EU32 A0A3B3I8F1 A0A2B4S341 A0A2S2PKE0 A0A131XQS8 L7MAD0 A0A2B4RF95 A0A147BB29 A0A1E1X2P5 A0A3B3HI99 A0A0P4VX79 A0A131XQR9 A0A147BJ69 A0A147BBR8 A0A1E1XGC7 A0A2H9T2V7 A0A0K8RPQ6 A0A131Y4G6 A0A131XV56 W4ZFN2 A0A131XRE4 A0A2G8JHC3 A0A147BL45 U5EQW9 A0A2H9T462 A0A2G8LLY0 A0A2H9T2U6 U5EQG0 A0A2G8KET5 A0A3B1IHU1 A0A2B4RTL1 V5H8T0 A0A147BID3 A0A3Q0IWM1 A0A2H9T361 A0A2B4SNL1 A0A147BI04 V5GPT7 A0A2H9T4F1 A0A2H9T3F5 A0A1E1XNX8 A0A147BLD9 W4Y3F1 A0A131XWB2 A0A146PGB4 A0A147BC70 A0A293LCV4 A0A147BK71 A0A146T2W5 A0A146QVS2 A0A182GL77 A0A131XWC9 A0A0P4VPK5 A0A3B3HKX5 A0A131XKD2 A0A0X3PQL9 A0A0X3PL94 A0A147BM11 Q4QQE6 T1DG92 A0A147BNC9 A0A3B1IHX1
Pubmed
EMBL
AY359886
AAQ57129.1
FM995623
CAX36787.1
ODYU01002162
SOQ39302.1
+ More
GDRN01105059 JAI57791.1 DPOC01000269 HCX22453.1 GFAC01000919 JAT98269.1 GEFM01002394 JAP73402.1 GEGO01001118 JAR94286.1 GEFM01005904 JAP69892.1 NSIT01000637 PJE77413.1 AAGJ04119271 FLMD02000280 SCV66640.1 GFAA01002514 JAU00921.1 GEGO01002473 JAR92931.1 ODYU01001756 SOQ38395.1 GBIH01002577 JAC92133.1 GEGO01004132 JAR91272.1 GEGO01004056 JAR91348.1 HAAD01000593 CDG66825.1 GFAC01005646 JAT93542.1 GDRN01111334 JAI56796.1 NSIT01000538 PJE77518.1 GEGO01007341 JAR88063.1 LSMT01000248 PFX22193.1 GEFM01002324 JAP73472.1 NSIT01000477 PJE77602.1 LSMT01000115 PFX26931.1 GEGO01003312 JAR92092.1 LSMT01000057 PFX30021.1 GEGO01004433 JAR90971.1 GANO01002501 JAB57370.1 LSMT01000222 PFX22988.1 GGMR01016717 MBY29336.1 GEFM01006407 JAP69389.1 GACK01003803 JAA61231.1 LSMT01000646 PFX15469.1 GEGO01007438 JAR87966.1 GFAC01005665 JAT93523.1 GDRN01111326 JAI56798.1 GEFM01006417 JAP69379.1 GEGO01004630 JAR90774.1 GEGO01007187 JAR88217.1 GFAC01000861 JAT98327.1 NSIT01000532 PJE77527.1 GADI01001229 JAA72579.1 GEFM01002398 JAP73398.1 GEFM01006250 JAP69546.1 AAGJ04124300 GEFM01005882 JAP69914.1 MRZV01001974 PIK35151.1 GEGO01003896 JAR91508.1 GANO01004026 JAB55845.1 NSIT01000294 PJE78013.1 MRZV01000037 PIK61266.1 NSIT01000539 PJE77517.1 GANO01004296 JAB55575.1 MRZV01000639 PIK46517.1 LSMT01000297 PFX20951.1 GANP01010944 JAB73524.1 GEGO01004857 JAR90547.1 NSIT01000462 PJE77627.1 PFX29995.1 GEGO01005479 JAR89925.1 GALX01002327 JAB66139.1 NSIT01000271 PJE78081.1 NSIT01000412 PJE77717.1 GFAA01002470 JAU00965.1 GEGO01003794 JAR91610.1 AAGJ04090645 GEFM01005840 JAP69956.1 GCES01143686 JAQ42636.1 GEGO01007027 JAR88377.1 GFWV01000938 MAA25668.1 GEGO01004248 JAR91156.1 GCES01099194 JAQ87128.1 GCES01126385 JAQ59937.1 JXUM01071389 KQ562662 KXJ75371.1 GEFM01005905 JAP69891.1 GDRN01111278 JAI56803.1 GEFH01000988 JAP67593.1 GEEE01008982 JAP54243.1 GEEE01011145 JAP52080.1 GEGO01003567 JAR91837.1 BN000795 CAJ00238.1 GALA01001771 JAA93081.1 GEGO01003111 JAR92293.1
GDRN01105059 JAI57791.1 DPOC01000269 HCX22453.1 GFAC01000919 JAT98269.1 GEFM01002394 JAP73402.1 GEGO01001118 JAR94286.1 GEFM01005904 JAP69892.1 NSIT01000637 PJE77413.1 AAGJ04119271 FLMD02000280 SCV66640.1 GFAA01002514 JAU00921.1 GEGO01002473 JAR92931.1 ODYU01001756 SOQ38395.1 GBIH01002577 JAC92133.1 GEGO01004132 JAR91272.1 GEGO01004056 JAR91348.1 HAAD01000593 CDG66825.1 GFAC01005646 JAT93542.1 GDRN01111334 JAI56796.1 NSIT01000538 PJE77518.1 GEGO01007341 JAR88063.1 LSMT01000248 PFX22193.1 GEFM01002324 JAP73472.1 NSIT01000477 PJE77602.1 LSMT01000115 PFX26931.1 GEGO01003312 JAR92092.1 LSMT01000057 PFX30021.1 GEGO01004433 JAR90971.1 GANO01002501 JAB57370.1 LSMT01000222 PFX22988.1 GGMR01016717 MBY29336.1 GEFM01006407 JAP69389.1 GACK01003803 JAA61231.1 LSMT01000646 PFX15469.1 GEGO01007438 JAR87966.1 GFAC01005665 JAT93523.1 GDRN01111326 JAI56798.1 GEFM01006417 JAP69379.1 GEGO01004630 JAR90774.1 GEGO01007187 JAR88217.1 GFAC01000861 JAT98327.1 NSIT01000532 PJE77527.1 GADI01001229 JAA72579.1 GEFM01002398 JAP73398.1 GEFM01006250 JAP69546.1 AAGJ04124300 GEFM01005882 JAP69914.1 MRZV01001974 PIK35151.1 GEGO01003896 JAR91508.1 GANO01004026 JAB55845.1 NSIT01000294 PJE78013.1 MRZV01000037 PIK61266.1 NSIT01000539 PJE77517.1 GANO01004296 JAB55575.1 MRZV01000639 PIK46517.1 LSMT01000297 PFX20951.1 GANP01010944 JAB73524.1 GEGO01004857 JAR90547.1 NSIT01000462 PJE77627.1 PFX29995.1 GEGO01005479 JAR89925.1 GALX01002327 JAB66139.1 NSIT01000271 PJE78081.1 NSIT01000412 PJE77717.1 GFAA01002470 JAU00965.1 GEGO01003794 JAR91610.1 AAGJ04090645 GEFM01005840 JAP69956.1 GCES01143686 JAQ42636.1 GEGO01007027 JAR88377.1 GFWV01000938 MAA25668.1 GEGO01004248 JAR91156.1 GCES01099194 JAQ87128.1 GCES01126385 JAQ59937.1 JXUM01071389 KQ562662 KXJ75371.1 GEFM01005905 JAP69891.1 GDRN01111278 JAI56803.1 GEFH01000988 JAP67593.1 GEEE01008982 JAP54243.1 GEEE01011145 JAP52080.1 GEGO01003567 JAR91837.1 BN000795 CAJ00238.1 GALA01001771 JAA93081.1 GEGO01003111 JAR92293.1
Proteomes
Pfam
Interpro
IPR000477
RT_dom
+ More
IPR005135 Endo/exonuclease/phosphatase
IPR036691 Endo/exonu/phosph_ase_sf
IPR030223 Dispatched
IPR003392 Ptc/Disp
IPR000731 SSD
IPR019786 Zinc_finger_PHD-type_CS
IPR011011 Znf_FYVE_PHD
IPR019787 Znf_PHD-finger
IPR013083 Znf_RING/FYVE/PHD
IPR005049 STL-like
IPR006594 LisH
IPR013087 Znf_C2H2_type
IPR036236 Znf_C2H2_sf
IPR015947 PUA-like_sf
IPR036987 SRA-YDG_sf
IPR003105 SRA_YDG
IPR005135 Endo/exonuclease/phosphatase
IPR036691 Endo/exonu/phosph_ase_sf
IPR030223 Dispatched
IPR003392 Ptc/Disp
IPR000731 SSD
IPR019786 Zinc_finger_PHD-type_CS
IPR011011 Znf_FYVE_PHD
IPR019787 Znf_PHD-finger
IPR013083 Znf_RING/FYVE/PHD
IPR005049 STL-like
IPR006594 LisH
IPR013087 Znf_C2H2_type
IPR036236 Znf_C2H2_sf
IPR015947 PUA-like_sf
IPR036987 SRA-YDG_sf
IPR003105 SRA_YDG
Gene 3D
ProteinModelPortal
Q6UV17
B9WPT7
A0A2H1VFZ3
A0A0P4VXU0
A0A3D5S1K1
A0A1E1XGQ7
+ More
A0A131Y4I8 A0A147BU27 A0A131XU84 A0A2H9T2H3 W4XNV3 A0A1G4M5I2 A0A1E1XNL1 A0A147BQ97 A0A2H1VC59 A0A090XE26 A0A147BLD0 A0A147BKN8 T2M4I6 A0A1E1X2T1 A0A0P4VTV9 A0A2H9T2R7 A0A147BBC3 W4YCB2 A0A2B4RZI8 A0A131Y4Q4 A0A2H9T2Z5 A0A2B4SAL9 A0A147BNQ9 A0A2B4SJF4 A0A147BJQ0 U5EU32 A0A3B3I8F1 A0A2B4S341 A0A2S2PKE0 A0A131XQS8 L7MAD0 A0A2B4RF95 A0A147BB29 A0A1E1X2P5 A0A3B3HI99 A0A0P4VX79 A0A131XQR9 A0A147BJ69 A0A147BBR8 A0A1E1XGC7 A0A2H9T2V7 A0A0K8RPQ6 A0A131Y4G6 A0A131XV56 W4ZFN2 A0A131XRE4 A0A2G8JHC3 A0A147BL45 U5EQW9 A0A2H9T462 A0A2G8LLY0 A0A2H9T2U6 U5EQG0 A0A2G8KET5 A0A3B1IHU1 A0A2B4RTL1 V5H8T0 A0A147BID3 A0A3Q0IWM1 A0A2H9T361 A0A2B4SNL1 A0A147BI04 V5GPT7 A0A2H9T4F1 A0A2H9T3F5 A0A1E1XNX8 A0A147BLD9 W4Y3F1 A0A131XWB2 A0A146PGB4 A0A147BC70 A0A293LCV4 A0A147BK71 A0A146T2W5 A0A146QVS2 A0A182GL77 A0A131XWC9 A0A0P4VPK5 A0A3B3HKX5 A0A131XKD2 A0A0X3PQL9 A0A0X3PL94 A0A147BM11 Q4QQE6 T1DG92 A0A147BNC9 A0A3B1IHX1
A0A131Y4I8 A0A147BU27 A0A131XU84 A0A2H9T2H3 W4XNV3 A0A1G4M5I2 A0A1E1XNL1 A0A147BQ97 A0A2H1VC59 A0A090XE26 A0A147BLD0 A0A147BKN8 T2M4I6 A0A1E1X2T1 A0A0P4VTV9 A0A2H9T2R7 A0A147BBC3 W4YCB2 A0A2B4RZI8 A0A131Y4Q4 A0A2H9T2Z5 A0A2B4SAL9 A0A147BNQ9 A0A2B4SJF4 A0A147BJQ0 U5EU32 A0A3B3I8F1 A0A2B4S341 A0A2S2PKE0 A0A131XQS8 L7MAD0 A0A2B4RF95 A0A147BB29 A0A1E1X2P5 A0A3B3HI99 A0A0P4VX79 A0A131XQR9 A0A147BJ69 A0A147BBR8 A0A1E1XGC7 A0A2H9T2V7 A0A0K8RPQ6 A0A131Y4G6 A0A131XV56 W4ZFN2 A0A131XRE4 A0A2G8JHC3 A0A147BL45 U5EQW9 A0A2H9T462 A0A2G8LLY0 A0A2H9T2U6 U5EQG0 A0A2G8KET5 A0A3B1IHU1 A0A2B4RTL1 V5H8T0 A0A147BID3 A0A3Q0IWM1 A0A2H9T361 A0A2B4SNL1 A0A147BI04 V5GPT7 A0A2H9T4F1 A0A2H9T3F5 A0A1E1XNX8 A0A147BLD9 W4Y3F1 A0A131XWB2 A0A146PGB4 A0A147BC70 A0A293LCV4 A0A147BK71 A0A146T2W5 A0A146QVS2 A0A182GL77 A0A131XWC9 A0A0P4VPK5 A0A3B3HKX5 A0A131XKD2 A0A0X3PQL9 A0A0X3PL94 A0A147BM11 Q4QQE6 T1DG92 A0A147BNC9 A0A3B1IHX1
Ontologies
PANTHER
Topology
Subcellular location
Nucleus
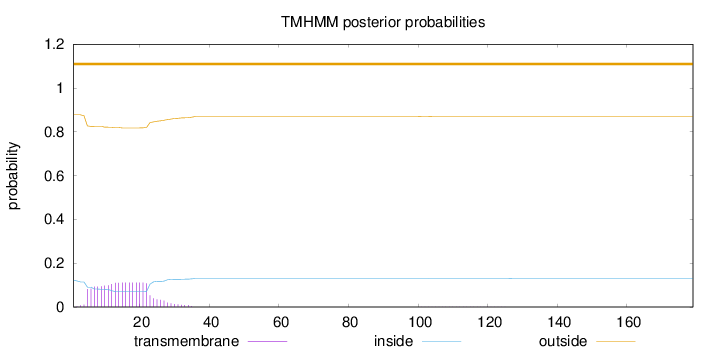
Length:
179
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.17593
Exp number, first 60 AAs:
2.16403
Total prob of N-in:
0.12235
outside
1 - 179
Population Genetic Test Statistics
Pi
56.139564
Theta
51.653084
Tajima's D
0.27036
CLR
0.110273
CSRT
0.444977751112444
Interpretation
Uncertain
