Gene
KWMTBOMO03496
Pre Gene Modal
BGIBMGA006346
Annotation
PREDICTED:_pre-mRNA-splicing_factor_18_isoform_X1_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 4.212
Sequence
CDS
ATGGATATTTTAAAGGCAGAAATTGCAAAAAAACGTAAATTACTTGAAGAAAAGAAAATTATTGATCCATCAAACAAGAAAAAATACTTCAAGAGAGGTGAGCTCTTAGCGAAAGAACAAGAAGAATATTTACGGAAATATGGACCCAAAGTACAGGAGCCTGAAGAAAAACAACTTGAAAAGGTAGAAACAGATCAAAATTATGCGAATGTAGAAACAGAACAGAAAGAGACTCCAGCACTTCCTCGAAATGAAGTTATAAAGAGATTGAGAGAAAGAGGTCATCCAATACTATTGTTTGCTGAAACTGAAATTCAGTCCTTTAAGAGATTAAGAAGAATTGAAATTCAAGAACCAGAAGTTAACAGGGGCTTCCGGAATGACTTCCAAGAAGCGATGGAGAAAGTCGATGAAGCATATTTGAATGAAATATTGGCATTAGGAACTCAGAATGAAGGTGAAAATTCGTCCAAAGAAGATGCACTTGATGATTCTATTACATATGAAACCATTCAAGAAATGGCAAAAACAATGGGACAAGGCGATAGAAACCATGATATGAATGTAATCATGACATTATTGCAATTCTTGCTAAAGTTGTGGGGTCAGCAACTGAGTTCTGCTGCAGCAGGACAGAAAGCAGCAATCAAGCATAAAATGACGAGAGCCACATACACCCAAACACAGGTTTATCTCAAGCCTCTAATGAGAAAACTAAAAAAGAAAAACTTGCCTGAAGACATCTGTGACAGTTTAACAGAAATAACAAAACATCTGTTAGATAGAAACTACATTATGGCCAGTGACGCATATCTTCAAATGGCGATTGGCAATGCGCCATGGCCAATCGGTGTAACCATGGTGGGTATTCACGCCCGTACAGGACGGGAGAAAATCTTCTCTAAGAACGTAGCACACGTTATGAACGACGAAACTCAAAGAAAATACATTCAAGCGCTTAAACGTCTTATGACTAAATGTCAGGAGTATTTCCCTACAGATCCATCTAGATGTGTGGAGTATACTACTATCAGATAA
Protein
MDILKAEIAKKRKLLEEKKIIDPSNKKKYFKRGELLAKEQEEYLRKYGPKVQEPEEKQLEKVETDQNYANVETEQKETPALPRNEVIKRLRERGHPILLFAETEIQSFKRLRRIEIQEPEVNRGFRNDFQEAMEKVDEAYLNEILALGTQNEGENSSKEDALDDSITYETIQEMAKTMGQGDRNHDMNVIMTLLQFLLKLWGQQLSSAAAGQKAAIKHKMTRATYTQTQVYLKPLMRKLKKKNLPEDICDSLTEITKHLLDRNYIMASDAYLQMAIGNAPWPIGVTMVGIHARTGREKIFSKNVAHVMNDETQRKYIQALKRLMTKCQEYFPTDPSRCVEYTTIR
Summary
Similarity
Belongs to the MCM family.
Uniprot
H9JA01
A0A2W1BU57
A0A2A4JLG4
A0A2H1WCP4
A0A212EIW6
S4NX41
+ More
A0A1E1VZ01 A0A194QBS7 A0A2J7QRC7 A0A0L7QPF7 A0A182GF53 A0A182XLZ9 Q16YV2 A0A182J483 A0A182VNQ8 A0A182UG31 Q7QBT9 A0A182HTG9 U5EY01 A0A182M9F0 A0A182XVF0 A0A023F847 A0A0V0GB36 A0A182FQ09 A0A1Q3FD33 A0A1B6KUY8 W5JIE6 A0A194QXS1 A0A1I8JUH1 A0A182QXV9 B0WCY0 A0A1B6IJF3 A0A2A3E2V7 A0A182JZB5 A0A310S8N7 A0A182W2M7 A0A232F4Z0 A0A1B0GJH8 R4FNF6 A0A182PEL5 A0A195FSE6 A0A1B6GTP5 F4WFL4 A0A336MML1 A0A336MP69 A0A154PDK8 E0VE67 A0A182NG98 D6W9R9 E2BBC4 A0A195CSS1 A0A158NSH5 A0A0J7NLU6 A0A0L0CJ86 E2AAJ0 A0A067RCM6 A0A1B0G846 A0A1B0ACL3 K7J6Z5 A0A0C9RJG6 E9JDH8 W8C5Z6 A0A3B0K905 A0A1I8MWR7 A0A026WKU6 A0A1W4UMU9 Q29BT5 B4GPN8 A0A1A9VR30 A0A1A9X187 A0A1A9XI51 A0A1B0BWJ8 A0A151WI78 B4NHY7 B4PMR0 A0A034VR84 B4K7A7 A0A0K8VU41 B3P379 A0A0A1WWS2 B4QTV8 Q9V437 A0A1Y1MTR3 A0A151HYS9 B4LXA4 B4ILV4 A0A151J4B7 A0A0A9YUK0 B3LVB7 A0A182S696 A0A0M4ELB8 A0A2R7WBQ0 A0A1I8NVF4 B4JSG2 A0A2M4AYP9 A0A0P4VJR2 A0A0K8TS61 E9HEU7 A0A182LFN8 A0A162T311
A0A1E1VZ01 A0A194QBS7 A0A2J7QRC7 A0A0L7QPF7 A0A182GF53 A0A182XLZ9 Q16YV2 A0A182J483 A0A182VNQ8 A0A182UG31 Q7QBT9 A0A182HTG9 U5EY01 A0A182M9F0 A0A182XVF0 A0A023F847 A0A0V0GB36 A0A182FQ09 A0A1Q3FD33 A0A1B6KUY8 W5JIE6 A0A194QXS1 A0A1I8JUH1 A0A182QXV9 B0WCY0 A0A1B6IJF3 A0A2A3E2V7 A0A182JZB5 A0A310S8N7 A0A182W2M7 A0A232F4Z0 A0A1B0GJH8 R4FNF6 A0A182PEL5 A0A195FSE6 A0A1B6GTP5 F4WFL4 A0A336MML1 A0A336MP69 A0A154PDK8 E0VE67 A0A182NG98 D6W9R9 E2BBC4 A0A195CSS1 A0A158NSH5 A0A0J7NLU6 A0A0L0CJ86 E2AAJ0 A0A067RCM6 A0A1B0G846 A0A1B0ACL3 K7J6Z5 A0A0C9RJG6 E9JDH8 W8C5Z6 A0A3B0K905 A0A1I8MWR7 A0A026WKU6 A0A1W4UMU9 Q29BT5 B4GPN8 A0A1A9VR30 A0A1A9X187 A0A1A9XI51 A0A1B0BWJ8 A0A151WI78 B4NHY7 B4PMR0 A0A034VR84 B4K7A7 A0A0K8VU41 B3P379 A0A0A1WWS2 B4QTV8 Q9V437 A0A1Y1MTR3 A0A151HYS9 B4LXA4 B4ILV4 A0A151J4B7 A0A0A9YUK0 B3LVB7 A0A182S696 A0A0M4ELB8 A0A2R7WBQ0 A0A1I8NVF4 B4JSG2 A0A2M4AYP9 A0A0P4VJR2 A0A0K8TS61 E9HEU7 A0A182LFN8 A0A162T311
Pubmed
19121390
28756777
22118469
23622113
26354079
26483478
+ More
17510324 12364791 14747013 17210077 25244985 25474469 20920257 23761445 28648823 21719571 20566863 18362917 19820115 20798317 21347285 26108605 24845553 20075255 21282665 24495485 25315136 24508170 30249741 15632085 17994087 17550304 25348373 25830018 10858562 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 28004739 25401762 26823975 27129103 26369729 21292972 20966253
17510324 12364791 14747013 17210077 25244985 25474469 20920257 23761445 28648823 21719571 20566863 18362917 19820115 20798317 21347285 26108605 24845553 20075255 21282665 24495485 25315136 24508170 30249741 15632085 17994087 17550304 25348373 25830018 10858562 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 28004739 25401762 26823975 27129103 26369729 21292972 20966253
EMBL
BABH01009467
KZ149899
PZC78612.1
NWSH01001141
PCG72424.1
ODYU01007651
+ More
SOQ50612.1 AGBW02014560 OWR41432.1 GAIX01009224 JAA83336.1 GDQN01011104 JAT79950.1 KQ459249 KPJ02445.1 NEVH01011914 PNF31139.1 KQ414815 KOC60523.1 JXUM01059244 JXUM01059245 KQ562042 KXJ76832.1 CH477508 EAT39810.1 AAAB01008859 EAA07486.3 APCN01000528 GANO01000646 JAB59225.1 AXCM01000235 GBBI01001251 JAC17461.1 GECL01001538 JAP04586.1 GFDL01009643 JAV25402.1 GEBQ01024727 JAT15250.1 ADMH02001428 ETN62564.1 KQ461155 KPJ08376.1 AXCN02001627 DS231891 EDS43940.1 GECU01020661 JAS87045.1 KZ288416 PBC26028.1 KQ764027 OAD54631.1 NNAY01000931 OXU25851.1 AJWK01021758 AJWK01021759 GAHY01001314 JAA76196.1 KQ981276 KYN43510.1 GECZ01003961 JAS65808.1 GL888120 EGI66991.1 UFQT01001722 SSX31526.1 UFQS01001453 UFQT01001453 SSX10958.1 SSX30623.1 KQ434878 KZC09897.1 DS235088 EEB11673.1 KQ971312 EEZ98537.1 GL446990 EFN87005.1 KQ977329 KYN03572.1 ADTU01024931 LBMM01003504 KMQ93455.1 JRES01000321 KNC32315.1 GL438128 EFN69537.1 KK852593 KDR20675.1 CCAG010000187 GBYB01013397 JAG83164.1 GL771866 EFZ09245.1 GAMC01004604 GAMC01004602 JAC01952.1 OUUW01000007 SPP82549.1 KK107161 QOIP01000003 EZA56588.1 RLU25301.1 CM000070 EAL26912.1 CH479186 EDW39121.1 JXJN01021835 KQ983089 KYQ47569.1 CH964272 EDW83637.2 CM000160 EDW95980.1 GAKP01013156 GAKP01013154 JAC45796.1 CH933806 EDW14231.1 GDHF01009918 JAI42396.1 CH954181 EDV48531.1 GBXI01011449 GBXI01006663 GBXI01001127 JAD02843.1 JAD07629.1 JAD13165.1 CM000364 EDX12381.1 AF170829 AE014297 AY118327 AAD49741.1 AAF55627.1 AAM48356.1 GEZM01021241 JAV89063.1 KQ976719 KYM76751.1 CH940650 EDW66756.1 CH480892 EDW55430.1 KQ980159 KYN17430.1 GBHO01007725 GBRD01006618 GDHC01008713 JAG35879.1 JAG59203.1 JAQ09916.1 CH902617 EDV43649.1 CP012526 ALC46065.1 KK854585 PTY17124.1 CH916373 EDV94702.1 GGFK01012586 MBW45907.1 GDKW01002124 JAI54471.1 GDAI01000838 JAI16765.1 GL732631 EFX69731.1 LRGB01000014 KZS21857.1
SOQ50612.1 AGBW02014560 OWR41432.1 GAIX01009224 JAA83336.1 GDQN01011104 JAT79950.1 KQ459249 KPJ02445.1 NEVH01011914 PNF31139.1 KQ414815 KOC60523.1 JXUM01059244 JXUM01059245 KQ562042 KXJ76832.1 CH477508 EAT39810.1 AAAB01008859 EAA07486.3 APCN01000528 GANO01000646 JAB59225.1 AXCM01000235 GBBI01001251 JAC17461.1 GECL01001538 JAP04586.1 GFDL01009643 JAV25402.1 GEBQ01024727 JAT15250.1 ADMH02001428 ETN62564.1 KQ461155 KPJ08376.1 AXCN02001627 DS231891 EDS43940.1 GECU01020661 JAS87045.1 KZ288416 PBC26028.1 KQ764027 OAD54631.1 NNAY01000931 OXU25851.1 AJWK01021758 AJWK01021759 GAHY01001314 JAA76196.1 KQ981276 KYN43510.1 GECZ01003961 JAS65808.1 GL888120 EGI66991.1 UFQT01001722 SSX31526.1 UFQS01001453 UFQT01001453 SSX10958.1 SSX30623.1 KQ434878 KZC09897.1 DS235088 EEB11673.1 KQ971312 EEZ98537.1 GL446990 EFN87005.1 KQ977329 KYN03572.1 ADTU01024931 LBMM01003504 KMQ93455.1 JRES01000321 KNC32315.1 GL438128 EFN69537.1 KK852593 KDR20675.1 CCAG010000187 GBYB01013397 JAG83164.1 GL771866 EFZ09245.1 GAMC01004604 GAMC01004602 JAC01952.1 OUUW01000007 SPP82549.1 KK107161 QOIP01000003 EZA56588.1 RLU25301.1 CM000070 EAL26912.1 CH479186 EDW39121.1 JXJN01021835 KQ983089 KYQ47569.1 CH964272 EDW83637.2 CM000160 EDW95980.1 GAKP01013156 GAKP01013154 JAC45796.1 CH933806 EDW14231.1 GDHF01009918 JAI42396.1 CH954181 EDV48531.1 GBXI01011449 GBXI01006663 GBXI01001127 JAD02843.1 JAD07629.1 JAD13165.1 CM000364 EDX12381.1 AF170829 AE014297 AY118327 AAD49741.1 AAF55627.1 AAM48356.1 GEZM01021241 JAV89063.1 KQ976719 KYM76751.1 CH940650 EDW66756.1 CH480892 EDW55430.1 KQ980159 KYN17430.1 GBHO01007725 GBRD01006618 GDHC01008713 JAG35879.1 JAG59203.1 JAQ09916.1 CH902617 EDV43649.1 CP012526 ALC46065.1 KK854585 PTY17124.1 CH916373 EDV94702.1 GGFK01012586 MBW45907.1 GDKW01002124 JAI54471.1 GDAI01000838 JAI16765.1 GL732631 EFX69731.1 LRGB01000014 KZS21857.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000235965
UP000053825
+ More
UP000069940 UP000249989 UP000076407 UP000008820 UP000075880 UP000075903 UP000075902 UP000007062 UP000075840 UP000075883 UP000076408 UP000069272 UP000000673 UP000053240 UP000075900 UP000075886 UP000002320 UP000242457 UP000075881 UP000075920 UP000215335 UP000092461 UP000075885 UP000078541 UP000007755 UP000076502 UP000009046 UP000075884 UP000007266 UP000008237 UP000078542 UP000005205 UP000036403 UP000037069 UP000000311 UP000027135 UP000092444 UP000092445 UP000002358 UP000268350 UP000095301 UP000053097 UP000279307 UP000192221 UP000001819 UP000008744 UP000078200 UP000091820 UP000092443 UP000092460 UP000075809 UP000007798 UP000002282 UP000009192 UP000008711 UP000000304 UP000000803 UP000078540 UP000008792 UP000001292 UP000078492 UP000007801 UP000075901 UP000092553 UP000095300 UP000001070 UP000000305 UP000075882 UP000076858
UP000069940 UP000249989 UP000076407 UP000008820 UP000075880 UP000075903 UP000075902 UP000007062 UP000075840 UP000075883 UP000076408 UP000069272 UP000000673 UP000053240 UP000075900 UP000075886 UP000002320 UP000242457 UP000075881 UP000075920 UP000215335 UP000092461 UP000075885 UP000078541 UP000007755 UP000076502 UP000009046 UP000075884 UP000007266 UP000008237 UP000078542 UP000005205 UP000036403 UP000037069 UP000000311 UP000027135 UP000092444 UP000092445 UP000002358 UP000268350 UP000095301 UP000053097 UP000279307 UP000192221 UP000001819 UP000008744 UP000078200 UP000091820 UP000092443 UP000092460 UP000075809 UP000007798 UP000002282 UP000009192 UP000008711 UP000000304 UP000000803 UP000078540 UP000008792 UP000001292 UP000078492 UP000007801 UP000075901 UP000092553 UP000095300 UP000001070 UP000000305 UP000075882 UP000076858
Interpro
Gene 3D
ProteinModelPortal
H9JA01
A0A2W1BU57
A0A2A4JLG4
A0A2H1WCP4
A0A212EIW6
S4NX41
+ More
A0A1E1VZ01 A0A194QBS7 A0A2J7QRC7 A0A0L7QPF7 A0A182GF53 A0A182XLZ9 Q16YV2 A0A182J483 A0A182VNQ8 A0A182UG31 Q7QBT9 A0A182HTG9 U5EY01 A0A182M9F0 A0A182XVF0 A0A023F847 A0A0V0GB36 A0A182FQ09 A0A1Q3FD33 A0A1B6KUY8 W5JIE6 A0A194QXS1 A0A1I8JUH1 A0A182QXV9 B0WCY0 A0A1B6IJF3 A0A2A3E2V7 A0A182JZB5 A0A310S8N7 A0A182W2M7 A0A232F4Z0 A0A1B0GJH8 R4FNF6 A0A182PEL5 A0A195FSE6 A0A1B6GTP5 F4WFL4 A0A336MML1 A0A336MP69 A0A154PDK8 E0VE67 A0A182NG98 D6W9R9 E2BBC4 A0A195CSS1 A0A158NSH5 A0A0J7NLU6 A0A0L0CJ86 E2AAJ0 A0A067RCM6 A0A1B0G846 A0A1B0ACL3 K7J6Z5 A0A0C9RJG6 E9JDH8 W8C5Z6 A0A3B0K905 A0A1I8MWR7 A0A026WKU6 A0A1W4UMU9 Q29BT5 B4GPN8 A0A1A9VR30 A0A1A9X187 A0A1A9XI51 A0A1B0BWJ8 A0A151WI78 B4NHY7 B4PMR0 A0A034VR84 B4K7A7 A0A0K8VU41 B3P379 A0A0A1WWS2 B4QTV8 Q9V437 A0A1Y1MTR3 A0A151HYS9 B4LXA4 B4ILV4 A0A151J4B7 A0A0A9YUK0 B3LVB7 A0A182S696 A0A0M4ELB8 A0A2R7WBQ0 A0A1I8NVF4 B4JSG2 A0A2M4AYP9 A0A0P4VJR2 A0A0K8TS61 E9HEU7 A0A182LFN8 A0A162T311
A0A1E1VZ01 A0A194QBS7 A0A2J7QRC7 A0A0L7QPF7 A0A182GF53 A0A182XLZ9 Q16YV2 A0A182J483 A0A182VNQ8 A0A182UG31 Q7QBT9 A0A182HTG9 U5EY01 A0A182M9F0 A0A182XVF0 A0A023F847 A0A0V0GB36 A0A182FQ09 A0A1Q3FD33 A0A1B6KUY8 W5JIE6 A0A194QXS1 A0A1I8JUH1 A0A182QXV9 B0WCY0 A0A1B6IJF3 A0A2A3E2V7 A0A182JZB5 A0A310S8N7 A0A182W2M7 A0A232F4Z0 A0A1B0GJH8 R4FNF6 A0A182PEL5 A0A195FSE6 A0A1B6GTP5 F4WFL4 A0A336MML1 A0A336MP69 A0A154PDK8 E0VE67 A0A182NG98 D6W9R9 E2BBC4 A0A195CSS1 A0A158NSH5 A0A0J7NLU6 A0A0L0CJ86 E2AAJ0 A0A067RCM6 A0A1B0G846 A0A1B0ACL3 K7J6Z5 A0A0C9RJG6 E9JDH8 W8C5Z6 A0A3B0K905 A0A1I8MWR7 A0A026WKU6 A0A1W4UMU9 Q29BT5 B4GPN8 A0A1A9VR30 A0A1A9X187 A0A1A9XI51 A0A1B0BWJ8 A0A151WI78 B4NHY7 B4PMR0 A0A034VR84 B4K7A7 A0A0K8VU41 B3P379 A0A0A1WWS2 B4QTV8 Q9V437 A0A1Y1MTR3 A0A151HYS9 B4LXA4 B4ILV4 A0A151J4B7 A0A0A9YUK0 B3LVB7 A0A182S696 A0A0M4ELB8 A0A2R7WBQ0 A0A1I8NVF4 B4JSG2 A0A2M4AYP9 A0A0P4VJR2 A0A0K8TS61 E9HEU7 A0A182LFN8 A0A162T311
PDB
1DVK
E-value=2.40283e-11,
Score=165
Ontologies
GO
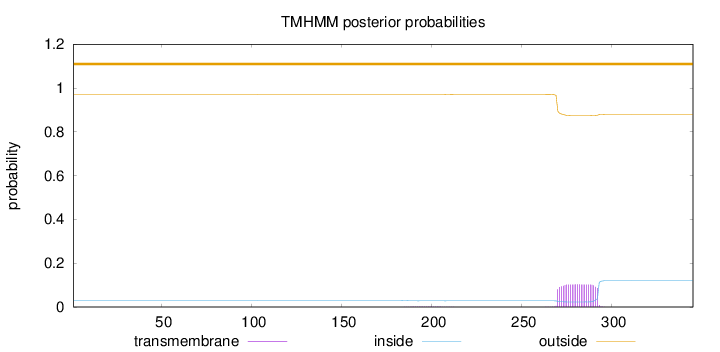
Topology
Length:
345
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.33709
Exp number, first 60 AAs:
0
Total prob of N-in:
0.02938
outside
1 - 345
Population Genetic Test Statistics
Pi
297.15288
Theta
202.820998
Tajima's D
1.472682
CLR
0.530619
CSRT
0.778611069446528
Interpretation
Uncertain
